Method
We have adopted the following steps for the identification of fusion transcripts in Paired-end RNA-seq datasets of Arabidopsis thaliana.
 Data download
Data download
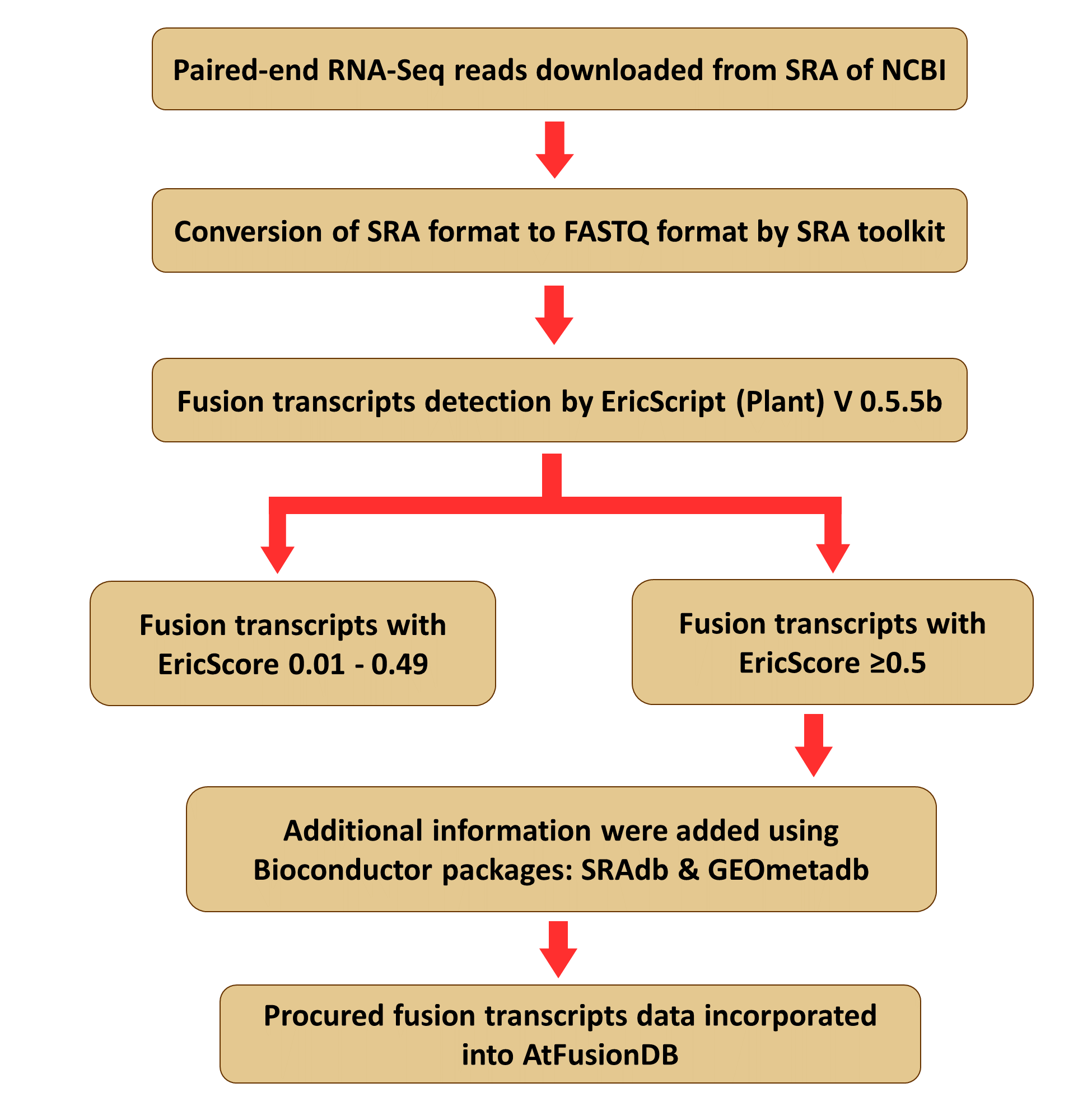
Paired-end RNA-Seq reads of Arabidopsis thaliana were downloaded from NCBI-SRA and converted into FASTQ file format using SRAtoolkit (version 2.8.2):
/PATH/TO/sratoolkit.2.8.2-1/bin/fastq-dump --split-3 <SRA_RUN-ID> -o </OUTPUT/FILE/PATH/ (default is present working directory)>  Running EricScript for fusion transcript detection
Running EricScript for fusion transcript detection
To list the available genomes at Ensembl Plants:
perl /PATH/TO/ERICSCRIPT-Plants/ericscript.pl --printdbAfter the selection of reference id (refid) (i.e. arabidopsis_thaliana), the Ensembl Database was downloaded (downdb) using the following command:
perl /PATH/TO/ERICSCRIPT-Plants/ericscript.pl --downdb --refid arabidopsis_thaliana -db </PATH/TO/DB_LOCATION> In order to perform fusion transcript detection from the samples of Arabidopsis thaliana, following command was executed using the default parameters.
By default, the database (-db) is taken from the following path: /PATH/TO/ERICSCRIPT-Plants/lib/data/arabidopsis_thaliana
perl /PATH/TO/ERICSCRIPT-Plants/ericscript.pl --refid <REFID> -name <SAMPLENAME> -o </PATH/TO/OUTPUT/> <R1.fastq> <R2.fastq> EricScript Output
EricScript Output
The /PATH/TO/OUTPUT/ folder contains the fusion transcripts detected in paired-end reads dataset. Identified fusion transcripts are reported in two files:
1. samplename.results.total.tsv: This file contains all the identified gene fusions.
2. samplename.results.filtered.tsv: This file contains the gene fusions with EricScore > 0.5
For all further processes we have finally considered the files having gene fusions with EricScore > 0.5 e.g. 'samplename.results.filtered.tsv'.
 Data preparation for AtFusionDB
Data preparation for AtFusionDB
Tissue type information of fusion transcripts was retrieved using the Bioconductor software package & SRA Run Selector. The utilities of Bioconductor package used are SRAdb and GEOmetadb. Combining all the results, the tissue information of the SRA samples was incorporated in AtFusionDB.
 Complete Pipeline
Complete Pipeline
The figure shown here is the repersentation of complete pipeline followed for the final data preparation of AtFusionDB.