Welcome to AtFusionDB
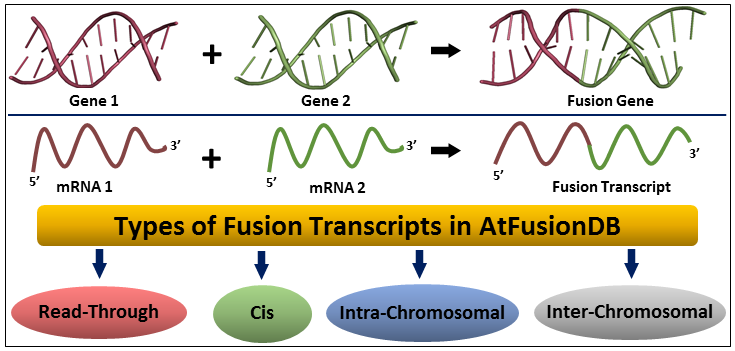
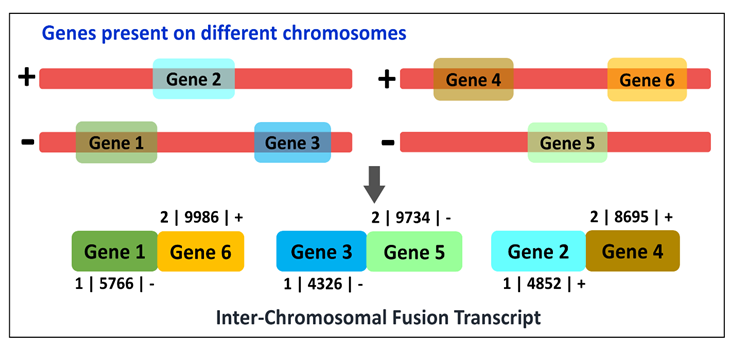
Fusion transcripts are chimeric mRNAs synthesized as a result of the fusion of two distinct genes or their transcripts. Till date, this domain has been explored largely in the animal kingdom, particularly in humans. However, some reports suggest that these chimeric transcripts may play a key role in altering overall cellular physiology in plants as well.
AtFusionDB is the first database dedicated to fusion transcripts in plants. It harbors 82,969 entries of different types of fusion transcripts identified in 3,520 paired-end RNA-Seq samples of the model plant Arabidopsis thaliana. This database is believed to be beneficial for exploring the complex and rarely explored domain of gene fusion and its implications in the plant kingdom.
Major Features of AtFusionDB
 Search
Search
Simple Search allows the user to search for any field and display any field (e.g. Fusion, Chromosome No., Gene-coordinates, SRA Id, Tissue and Junction-sequence related information). Advanced Search is a level up of the simple search option. It allows to search for specific fusion queries if the user has prehanded information. It is very efficient and time saving when the user is looking for a specific fusion transcript in AtFusionDB.
 Browse
Browse
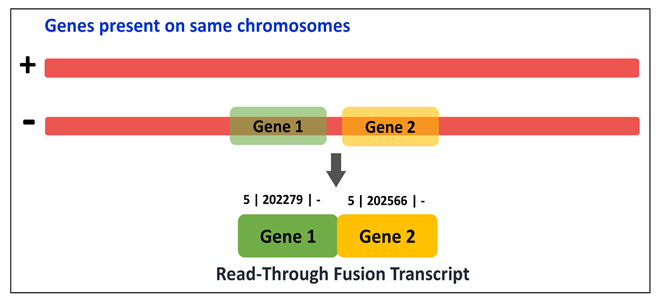
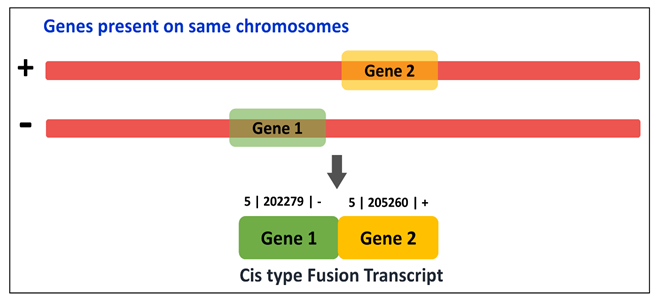
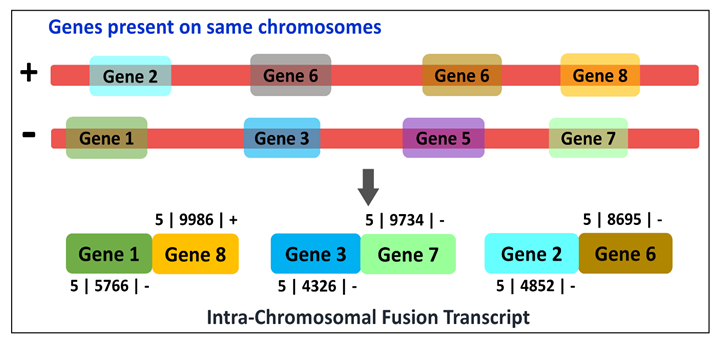
It can be utilized by four modes viz. Fusion type-wise classification, Tissue type-wise classification, EricScore-wise categorization and Chromosome-wise classification.
 Tools
Tools
Several web based tools have been integrated into AtFusionDB that includes Smith-Waterman Aligner (for alignment based on Smith-Waterman algorithm), Mapping (for mapping of fusion transcript junction sequences to user provided nucleotide sequences/Genomic DNA) and BLAST (for homology search).
 Other features
Other features
Uniprot-ID(s), EnsemblPlants Id, fused-gene expression, strand sense, individual gene function and their expression, fusion name, breakpoint information and SRA Id which will help the user to gain further information about fusion transcripts of Arabdiopsis thaliana.