Help
This page is designed to guide the users so that they can understand PlantPepDB database and use it effectively. Here users can get the detailed information about all the modules and how to use them to get the most out of PlantPepDB. The steps and the outcome of every step is explained in images with description. User can directly go to the particular section by clicking the respective menu present in the main table.
Search
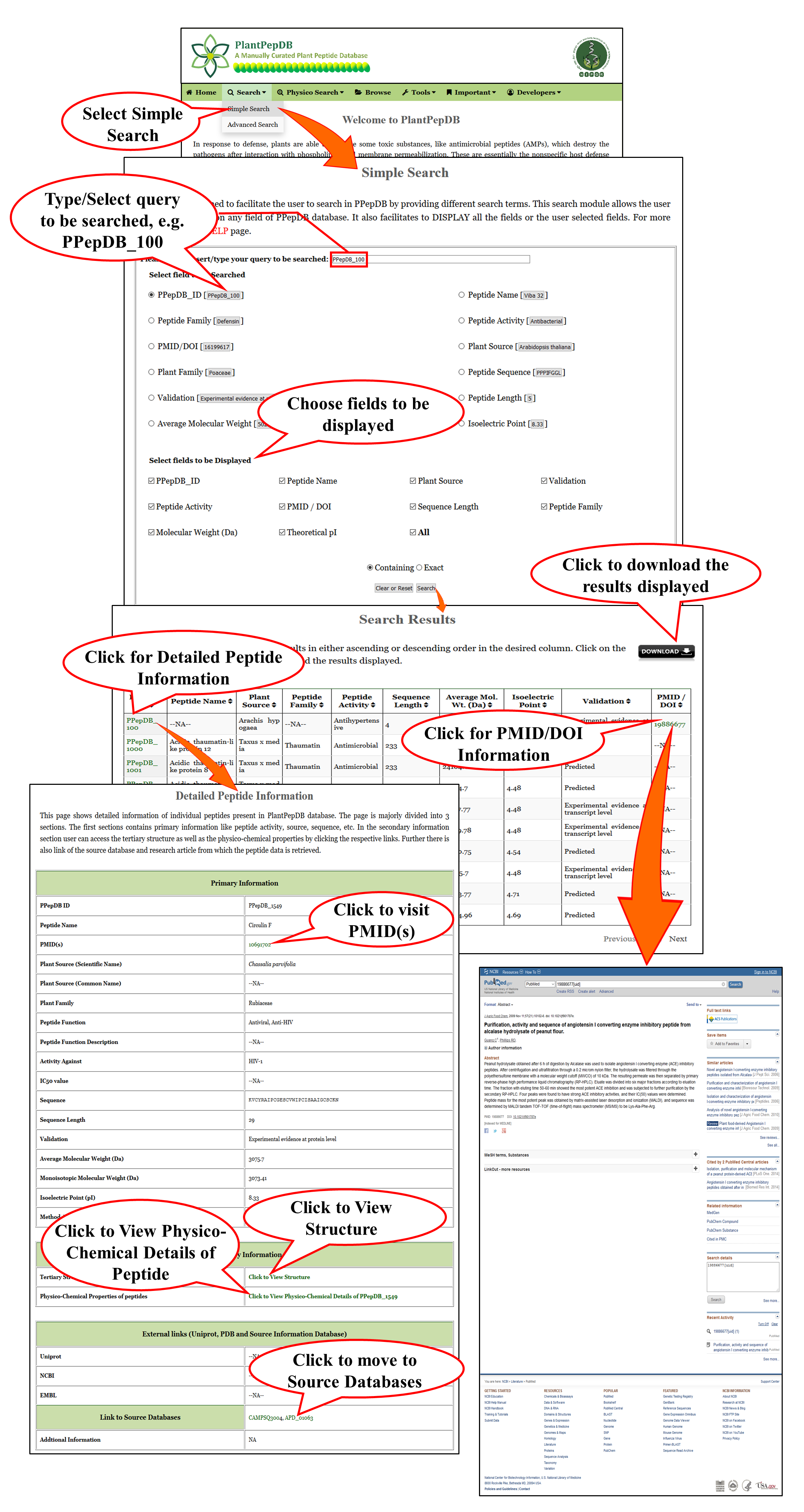
Simple Search |
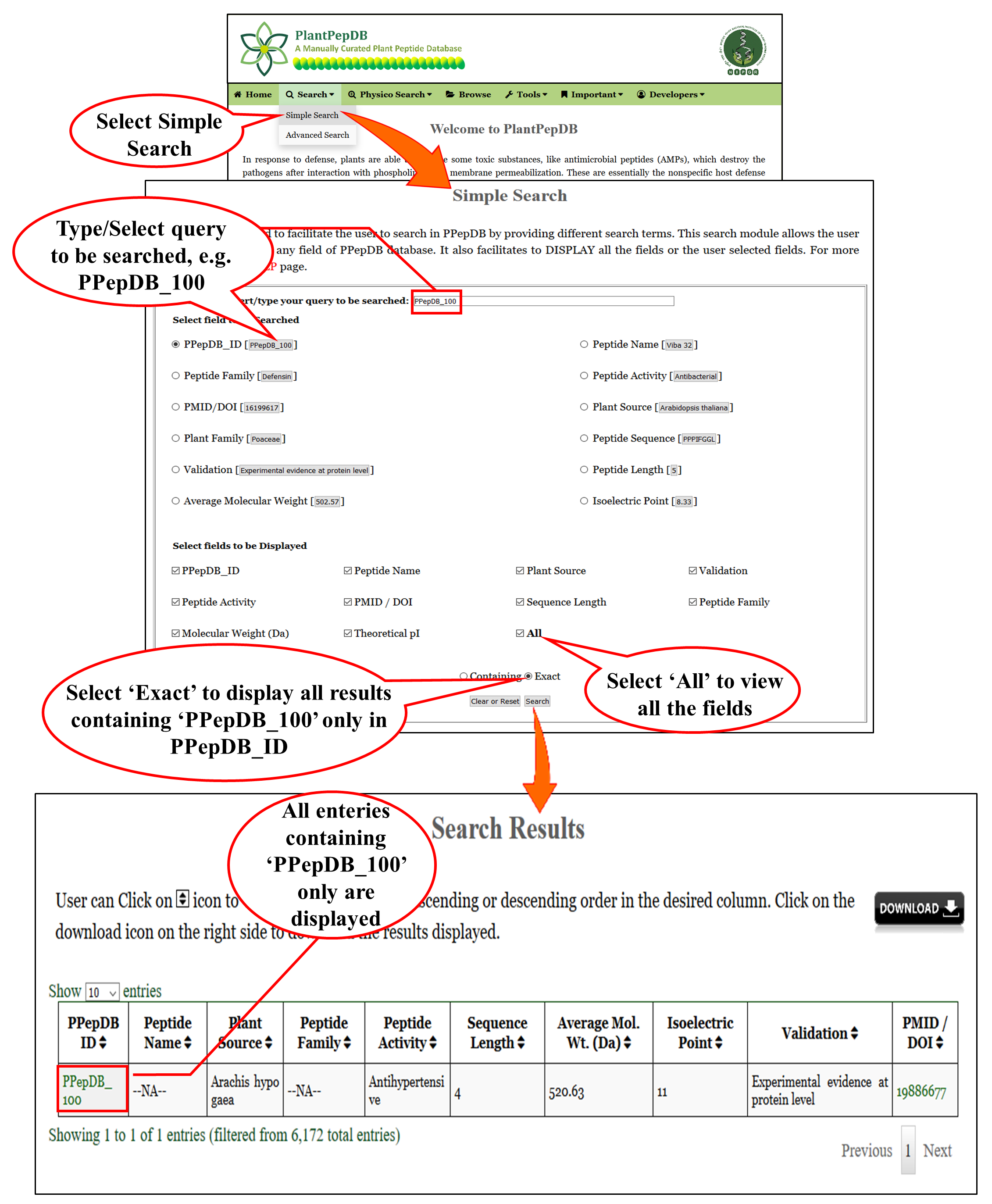
| This is a very basic and user friendly search option. User can search any query in any of the fields (e.g. PPepDB ID PPepDB_100) of the database. Results will be displayed according to the selected fields to be dispalyed. e.g. PPepDB ID, Peptide Name, Source, etc. |
1. Default Condition
2. When clicked on "Containing"
3. When clicked on "Exact"
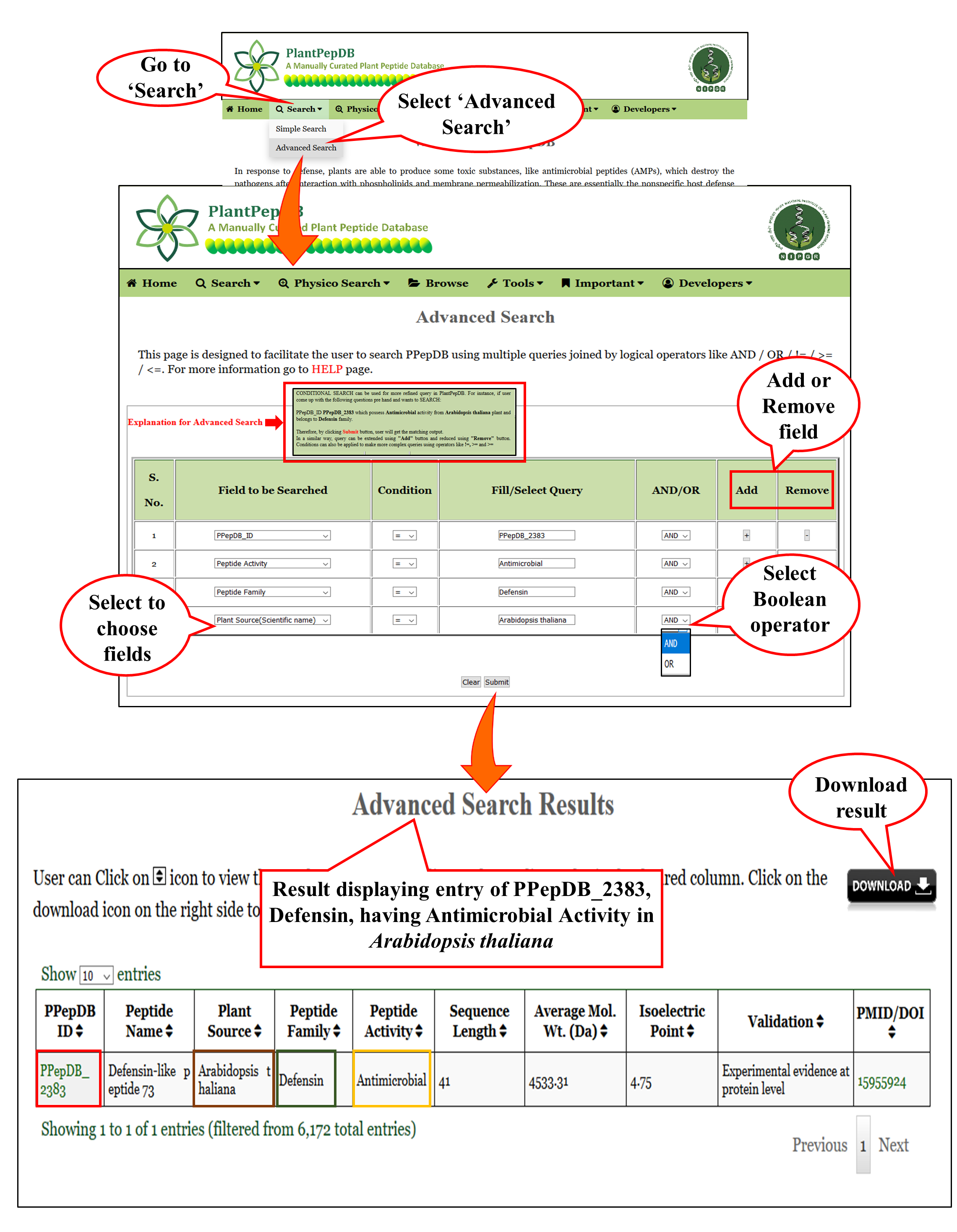
Advanced Search |
| This is a level-up from the usual simple search option. User can search PlantPepDB by using multiple queries combined with logical operators like AND / OR / != / > / < / =. All the data matching with the query, will be displayed with all other related information available in the database. |
Physico Search
AA Composition Search |
| This module allows the user to seach PlantPepDB based on sequence information. User have to provide amino acid compositon information of the peptide sequence by selecting the amino acids and their minimum & maximum values. |

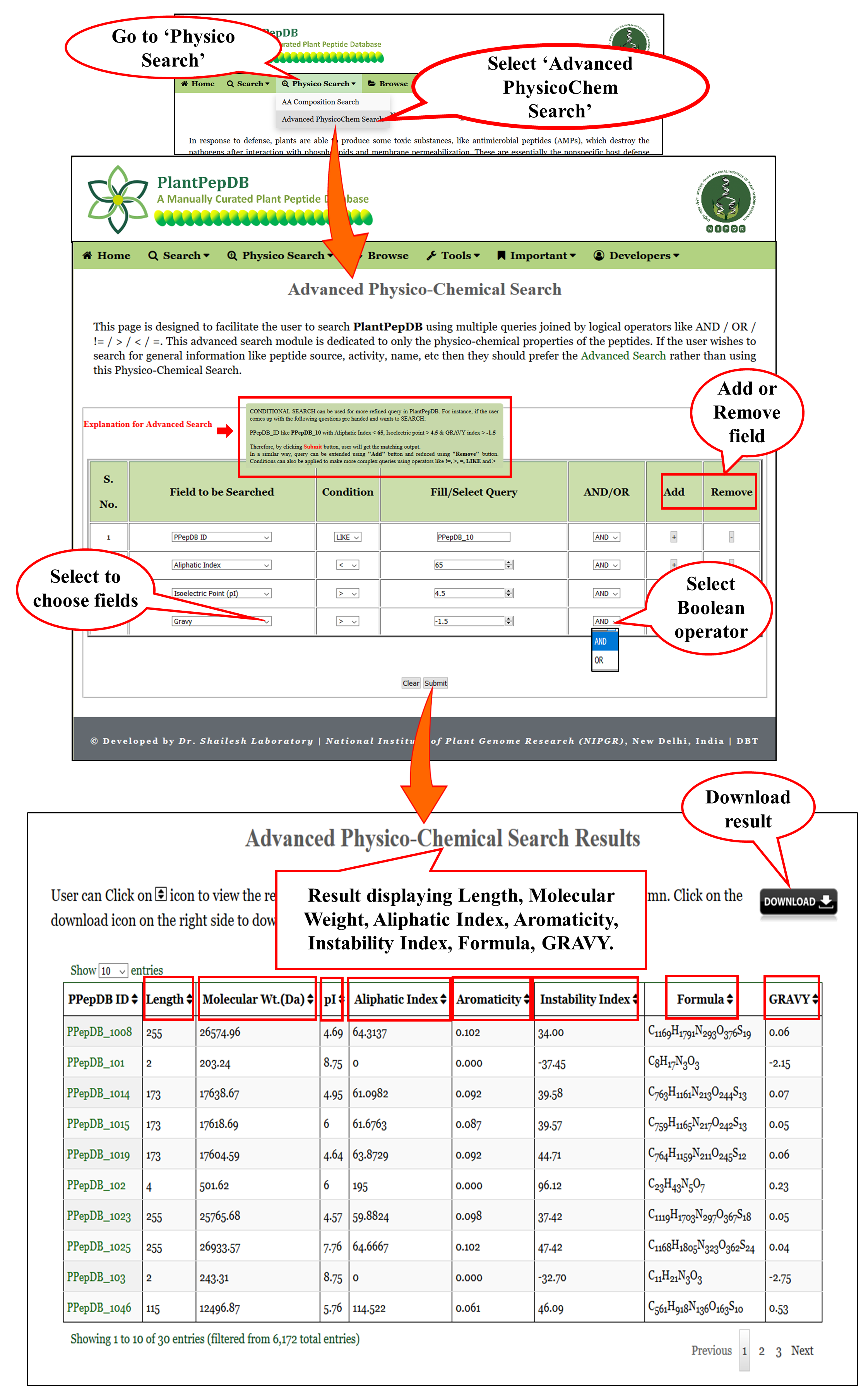
Advanced PhysicoChem Search |
| This module allows the user to search PlantPepDB based on various Physico-Chemical properties of the peptides. There are multiple operators in the search option which, the user can use to make complex queries. |
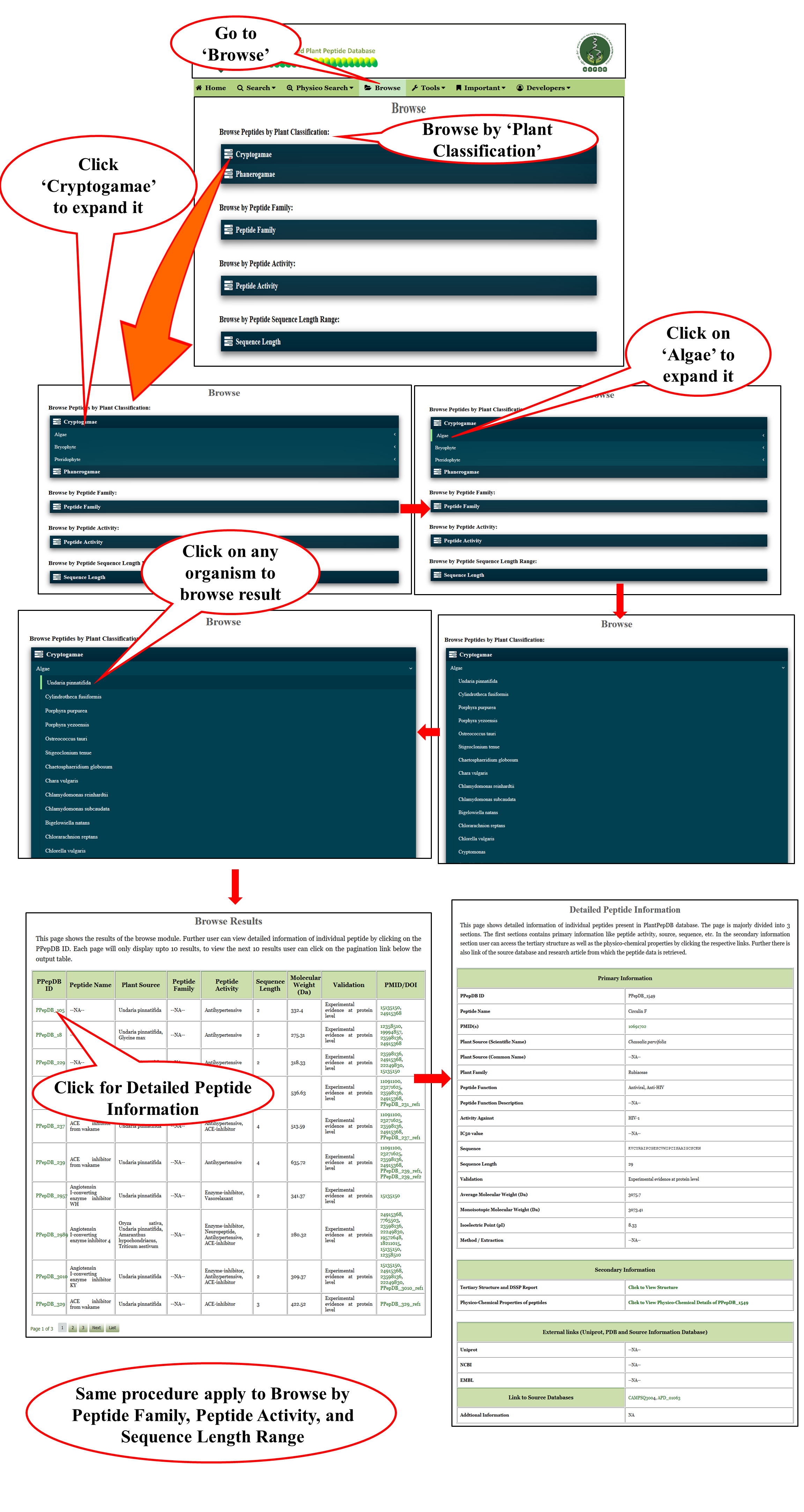
Browse PlantPepDB
Browse |
| The browse page is designed in a user friendly way. Overall it provides four different options to browse from i.e. plant source, peptide family, peptide activity and sequence length. |
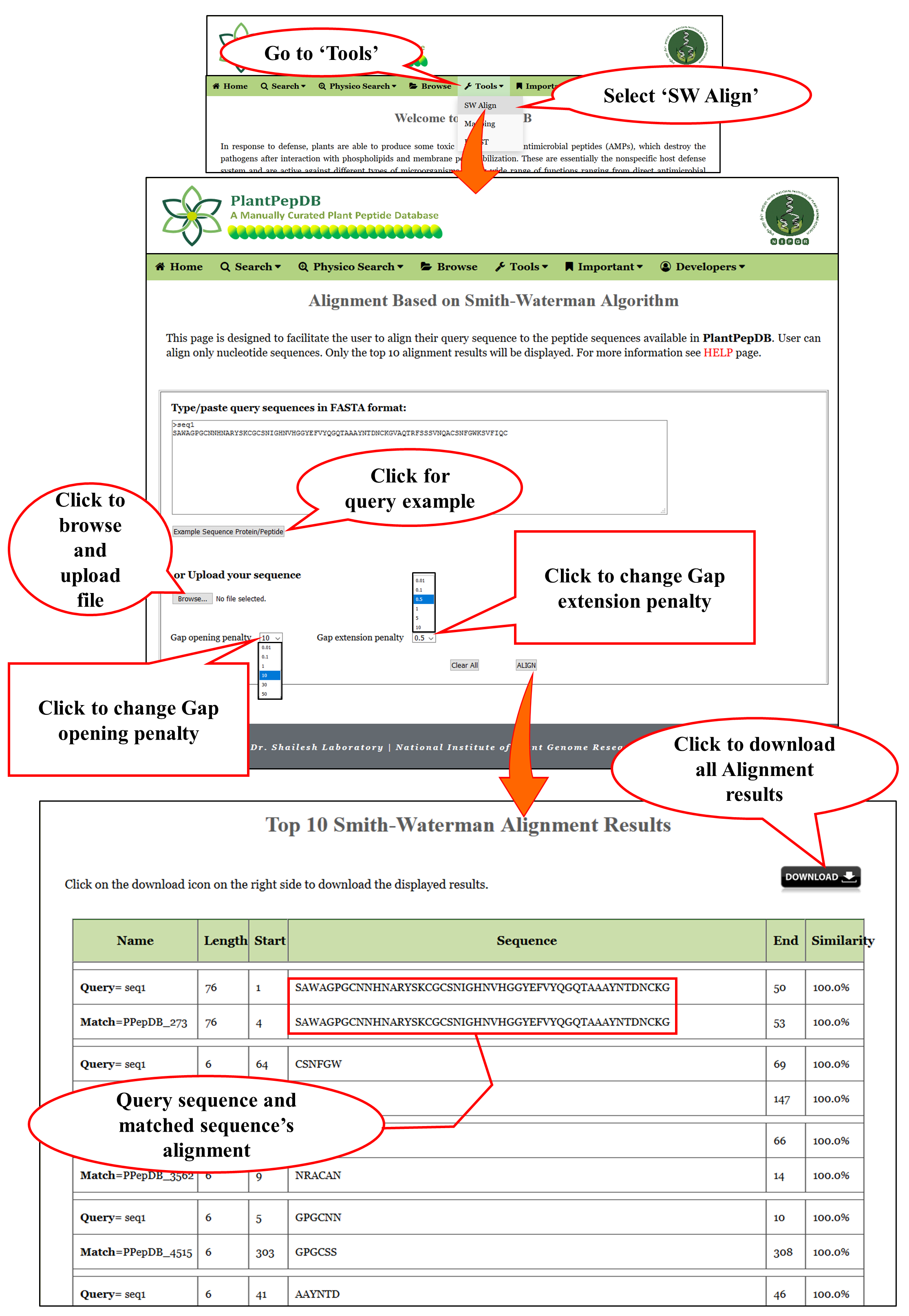
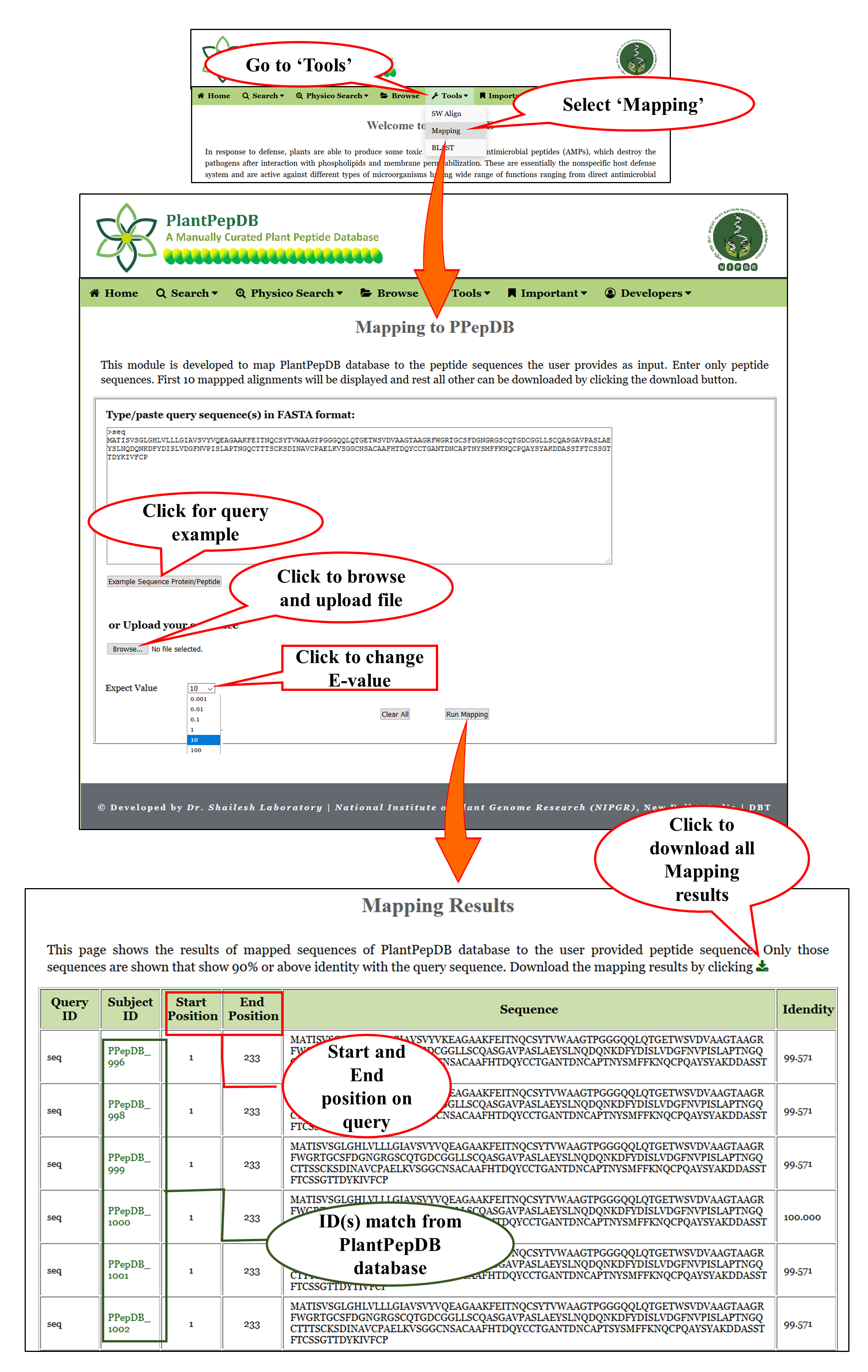
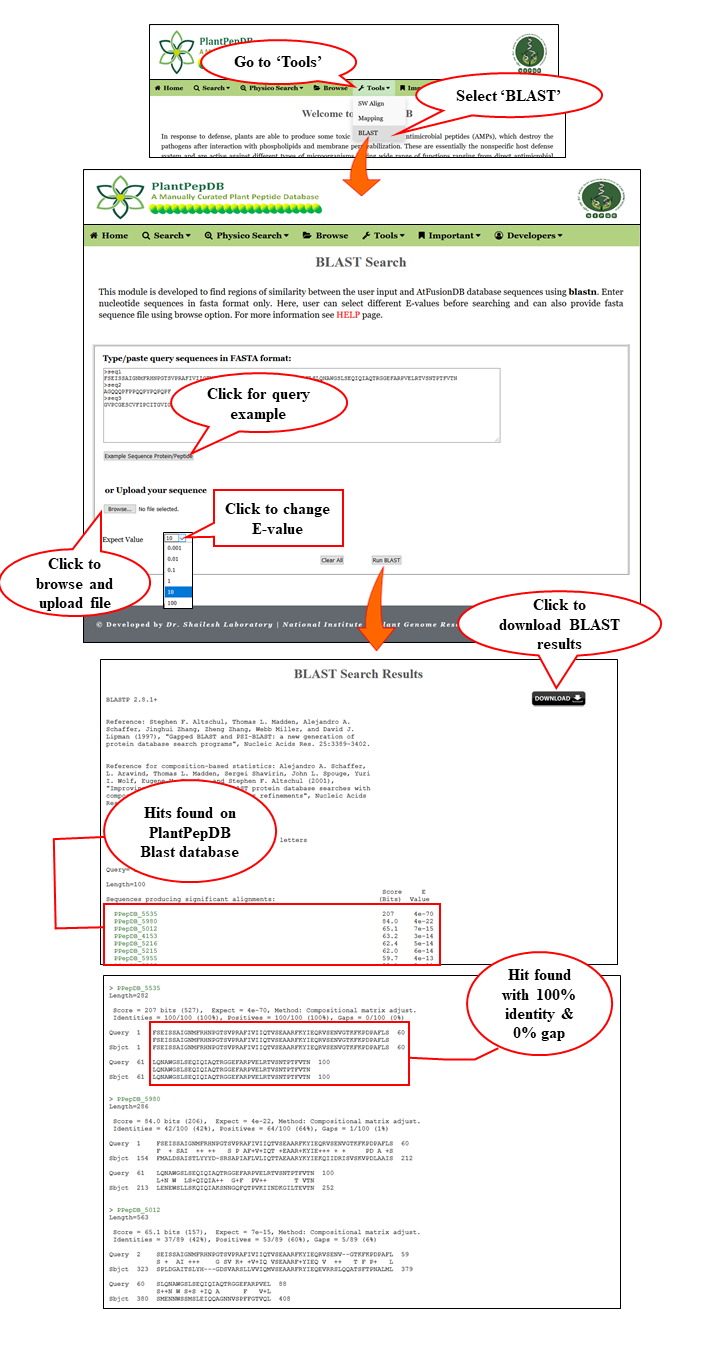
Tools available in PlantPepDB
SW Align |
| SW align i.e. Smith-Waterman Alignment page assists the user to run a Local alignment of a sequence using Smith-Waterman algorithm. User can align their query sequence to all the peptide sequences available in PlantPepDB. |
Mapping |
| This module maps the peptide sequences that are available in PlantPepDB on the user provided peptide sequence. Mapping can be done against all the sequences availabel in PlantPepDB. User can select different E-values before mapping. Default E-value is set at 10. |
BLAST |
| User can run a BLAST search for peptide sequences. The server provides hits against whole PlantPepDB database. Different E-values are provided, from which user can select the suitable one. Default E-value is set at 10. |
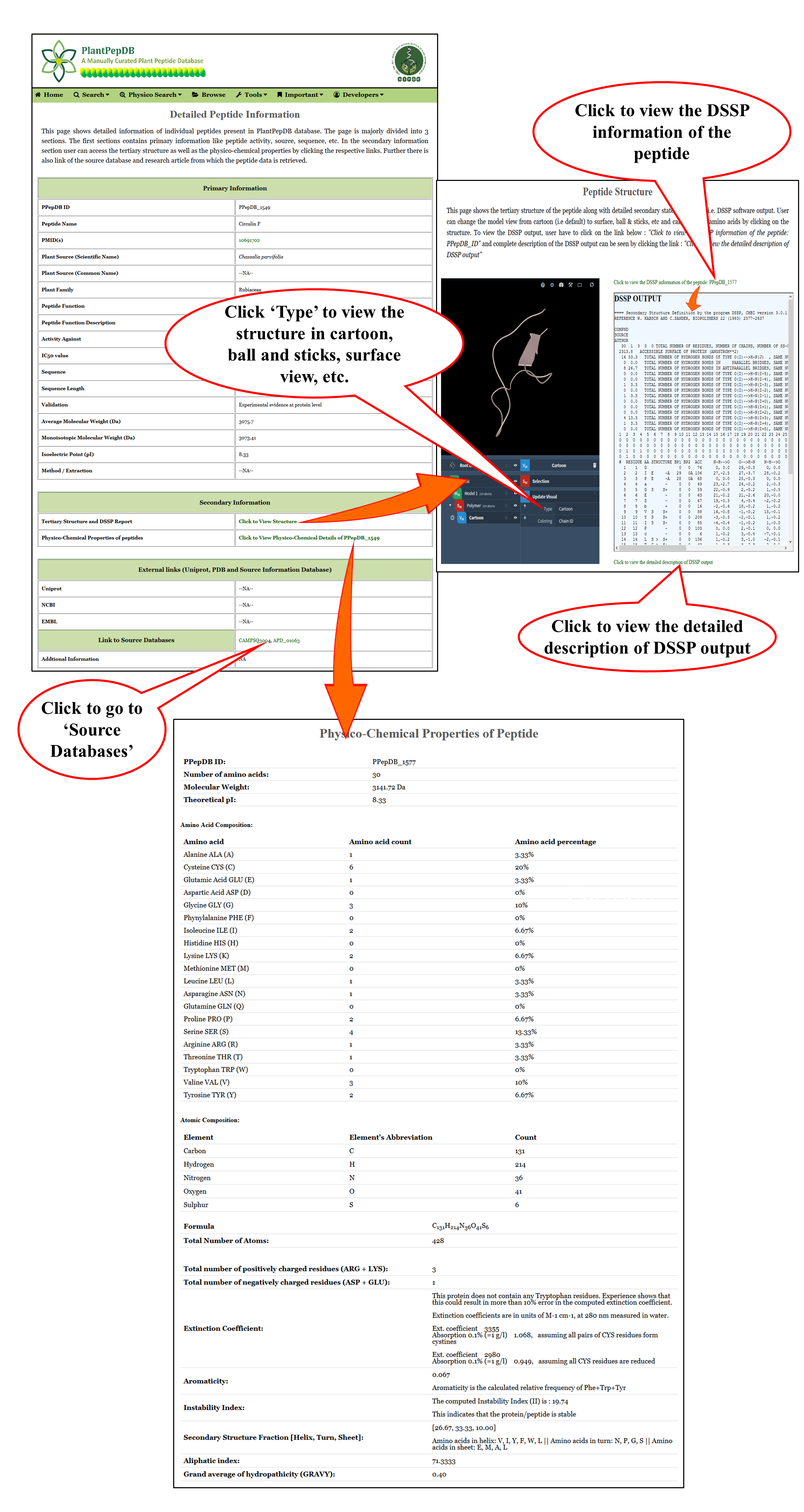
Structural & physico-chemical information |
| These are the pages in which user can view various details like basic peptide information, physico-chemical information, tertiary structure and DSSP state information of individual peptide entry present in PlantPepDB database. |