Method
Small RNA seq reads download
Total of 2434 sRNA-seq data [Arabidopsis thaliana (1676), Solanum lycopersicum (142), Cicer arietinum (25), Medicago truncatula (127), Oryza sativa (243), and Zea mays (221)], available at Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra), from the National Center for Biotechnology Information (NCBI) were downloaded.
Read processing and tRNA nucleoside modification sites prediction
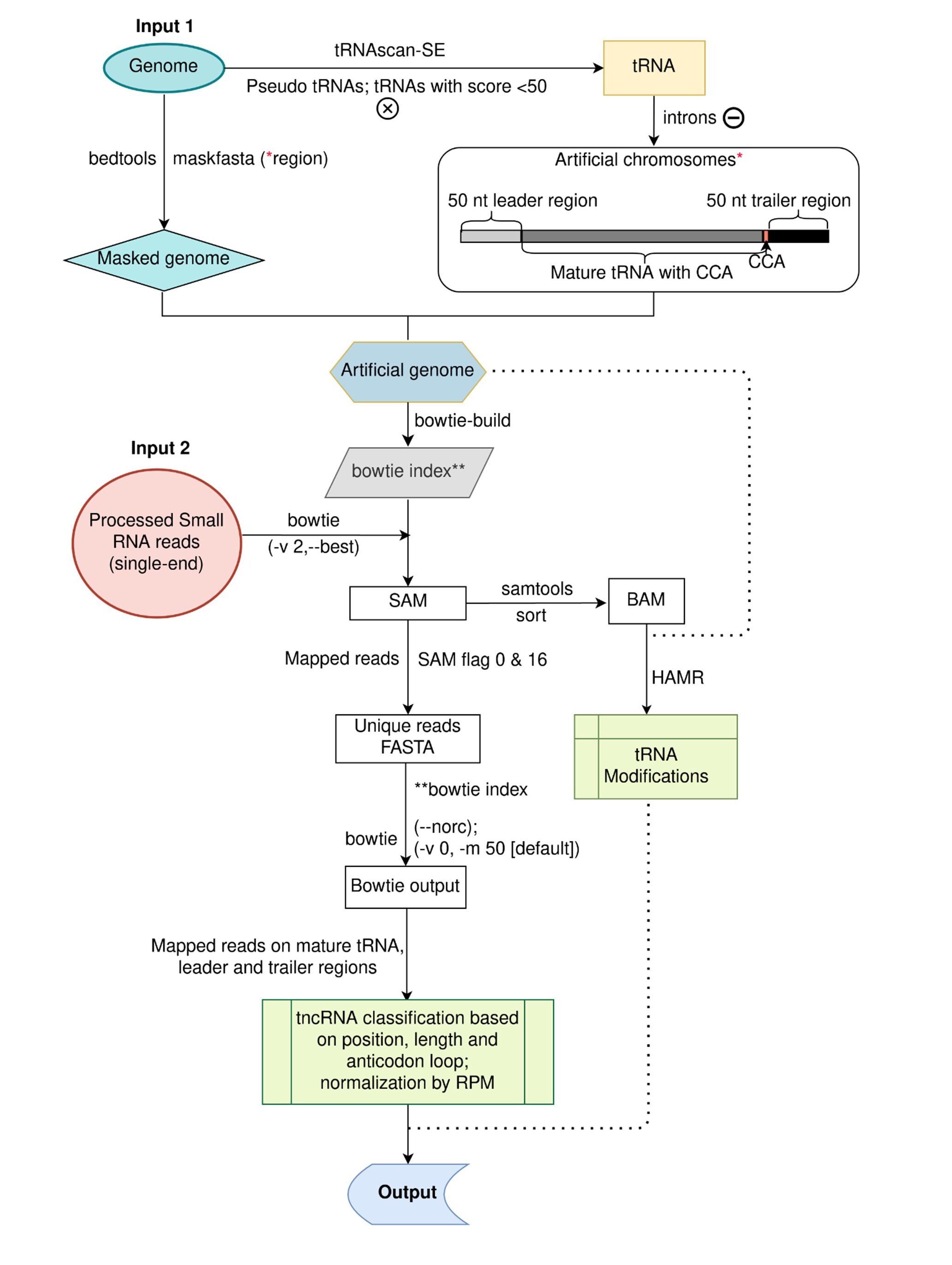
The single-end small RNA reads were processed by Trim Galore v0.6.6 (https://github.com/FelixKrueger/TrimGalore) to remove the low-quality bases and trim the adapter sequences. To permit error-tolerant and uniquely mapping reads alignments, processed reads were aligned to the artificial genome index using bowtie with ‘-v2 --best’ options. The SAM output was sorted and converted to BAM by samtools (v1.10).
The modification sites were predicted by HAMR (v1.2) (python hamr.py IN.bam REF.fa models/euk_trna_mods.Rdata OUT ath 30 10 0.05 H4 0.01 0.05 0.05).
Modification sites on the mature tRNA and leader/trailer regions were counted.
tncRNAs identification and classification
After processing and quality control, filtered reads were provided as input to the in-house pipeline. Due to the huge number of tRNA gene copies predicted by tRNAscanSE, we have filtered our tRNA gene pool by eliminating pseudogenes and keeping genuine tRNAs based on score. Only high-quality tRNAs with a score equal to or above 50 were selected for mapping. To avoid ambiguous reads from non-tRNA regions, we created an artificial genome by masking the genuine tRNA gene region with 50 nt upstream and downstream in the genome (masked genome) and adding them as artificial chromosomes. We classified only those reads as tncRNAs that mapped exclusively to the artificial chromosomes (i.e. tRNA set consisting of mature tRNA, 50 nt leader, and 50 nt trailer). The identified tncRNAs were categorized into tRF-5, tRF-3, tRF-1, leader-tRF, 5′tRH, 3′tRH, and other tRFs.
© Developed by Dr. Shailesh Laboratory | National Institute of Plant Genome Research (NIPGR), New Delhi, India | DBT
