Help
This page is designed to guide users about using PVsiRNAdb. It constitutes of various snapshots of various procedures, facilities and understading the output.
| Search | Browse | Tools |
|---|---|---|
| Simple Search | Browse by Virus Name | BLAST |
| Batch Search | Browse by Plant Name | SW Align |
| Browse by PubMed Id | Mapping |
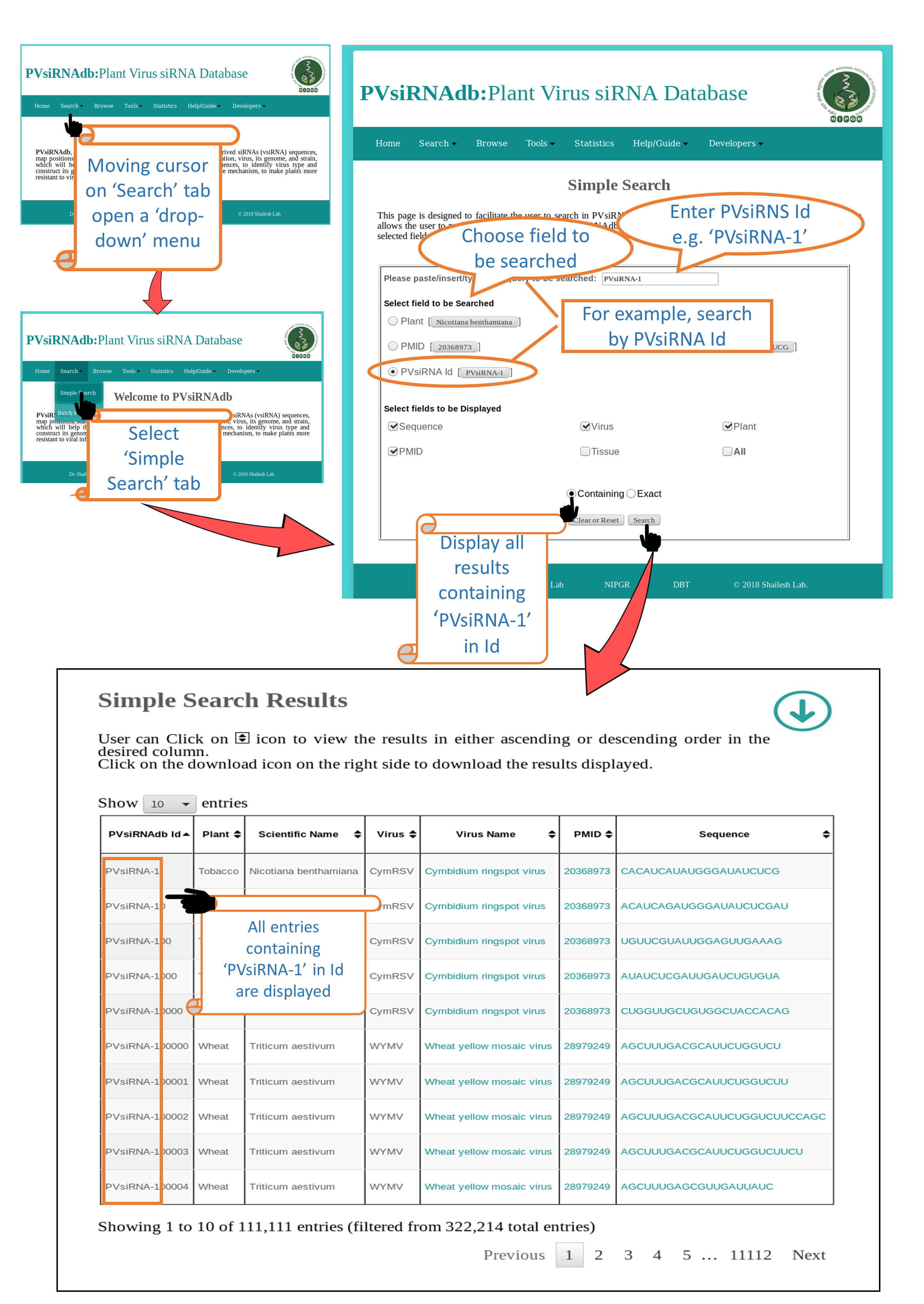
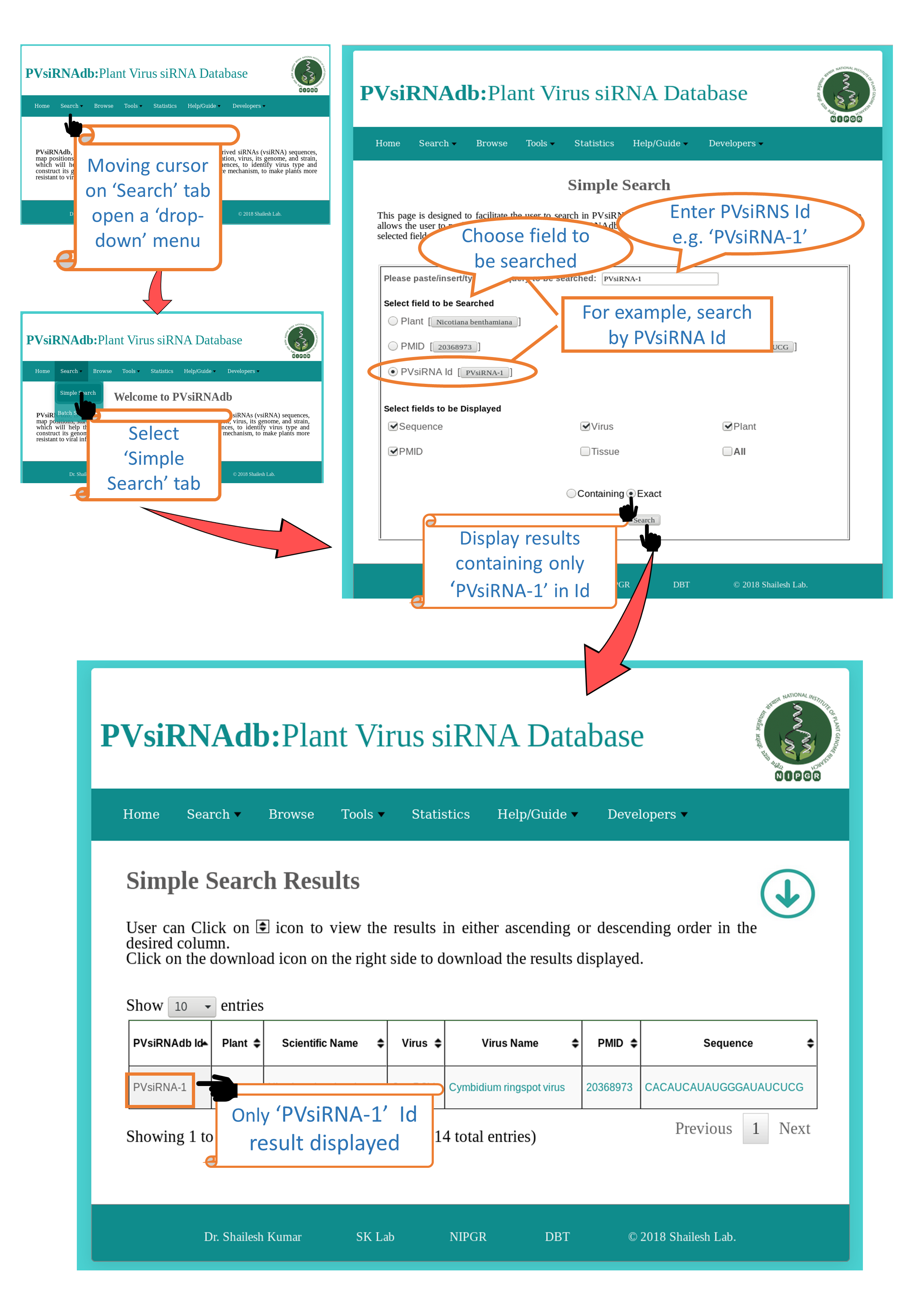
Search
Simple Search |
| This is a very simple and easy search option. User can search any query in any of the fields (e.g. Plant Nicotiana Benthamiana).Results will be displayed according to the selected fields to be dispalyed. e.g. Virus, Sequence, PMID etc. |
1. Default Condition

2. When clicked on "Containing"
3. When clicked on "Exact"
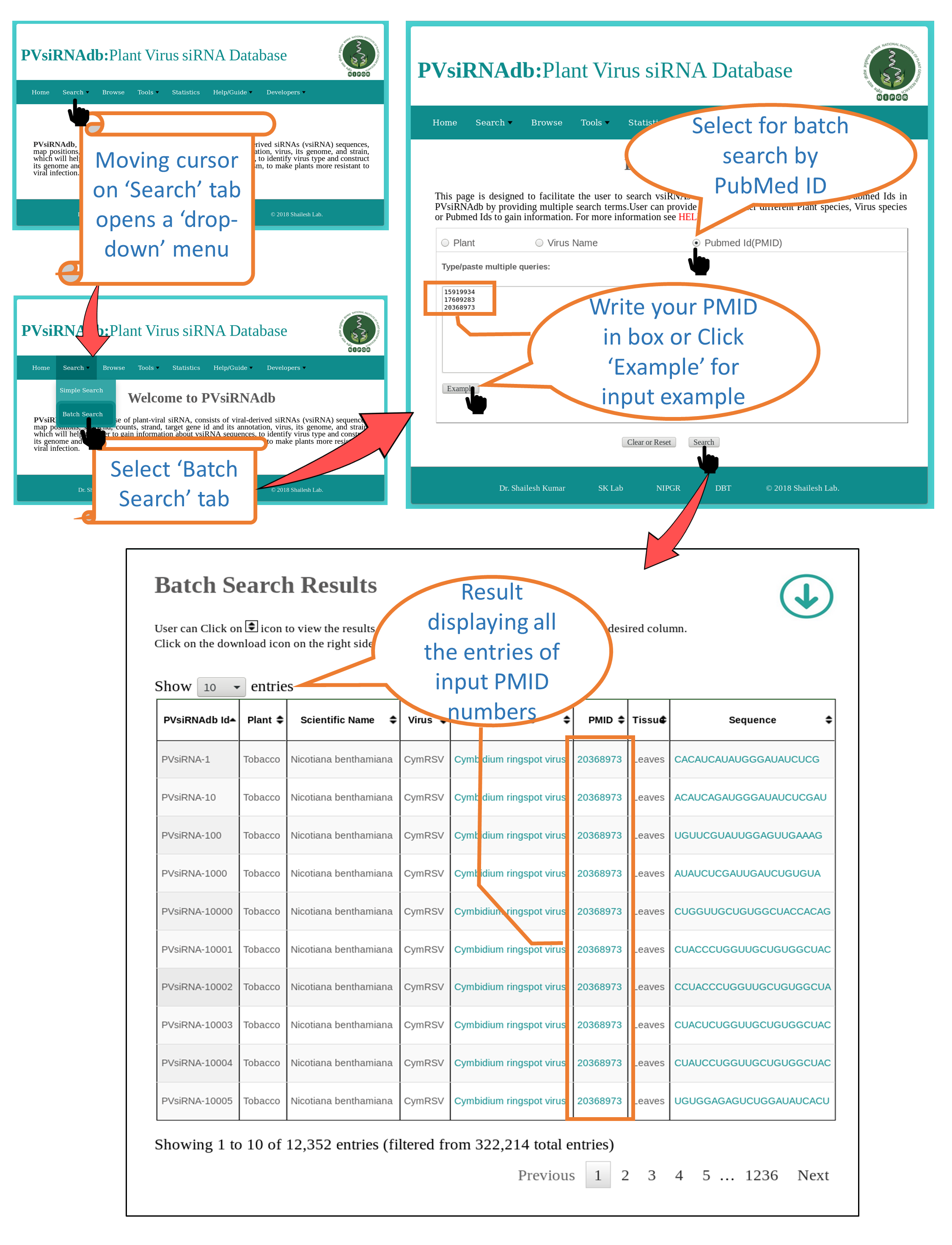
Batch Search |
| This is a level-up part of search option. User can search multiple queries in any of the fields (e.g. Plant , Virus and PubMed Id).All the results matching to the queries will be displayed with all the information available. |
1. Search by "Plant Name"

2. Search by "Virus Name"

3. Search by "PubMed Id
Browse
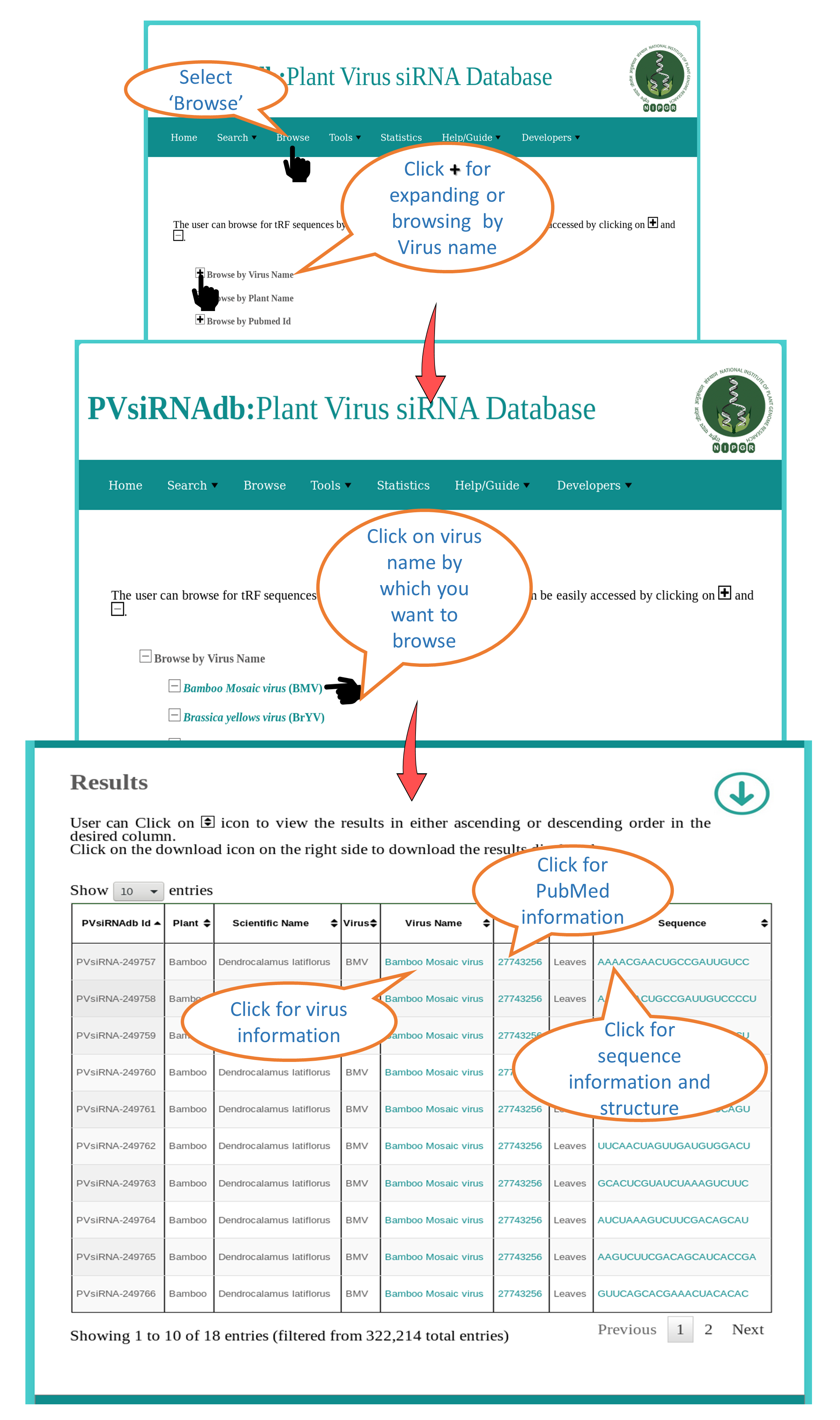
Browse by Virus Name |
| This interface allows the user to browse all the virus specific siRNAs available in PVsiRNAdb. All the available information will be dispalyed e.g. Virus, Sequence, PMID etc. |
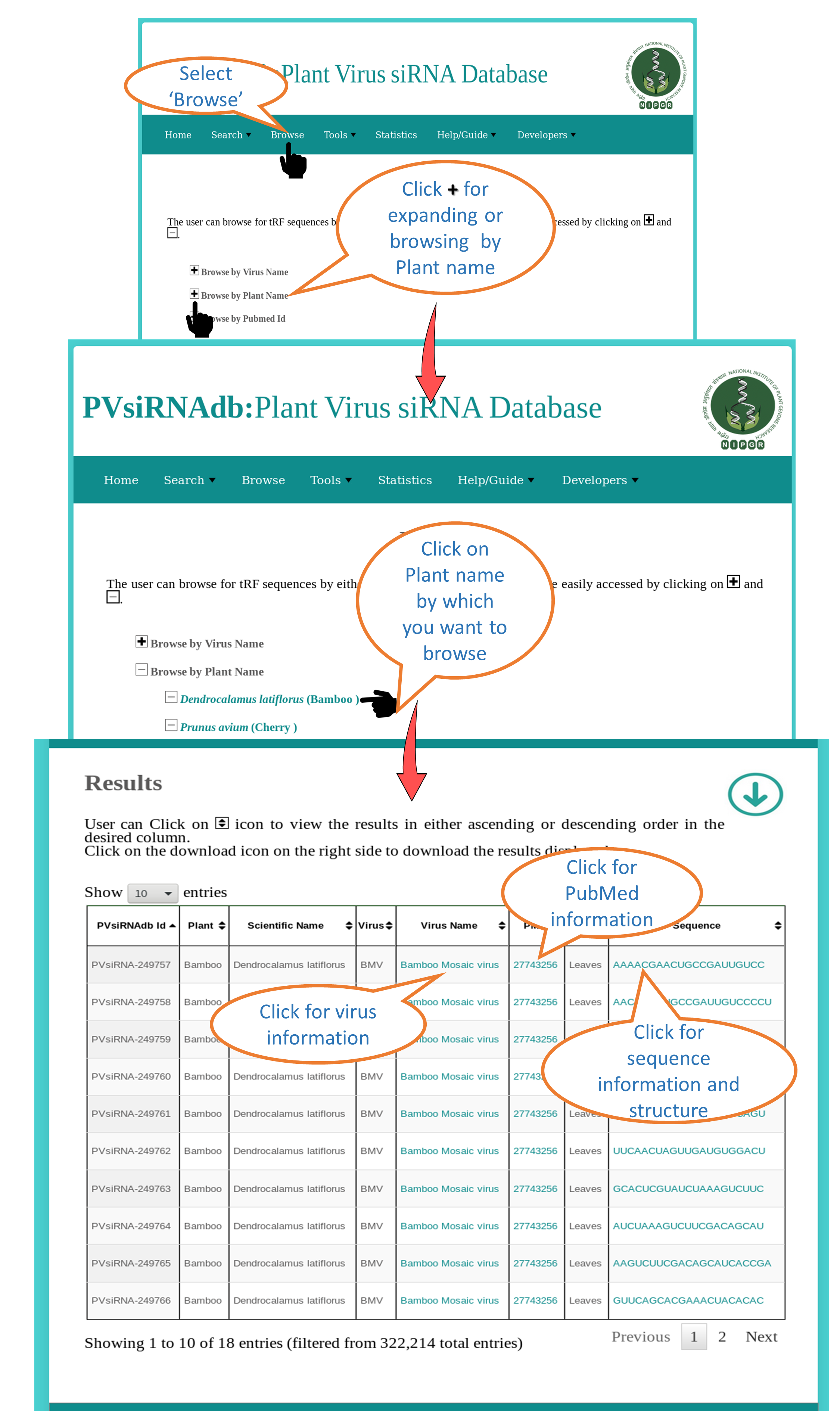
Browse by Plant Name |
| This interface allows the user to browse all the plant specific siRNAs available in PVsiRNAdb. All the available information will be dispalyed e.g. Virus, Sequence, PMID etc. |
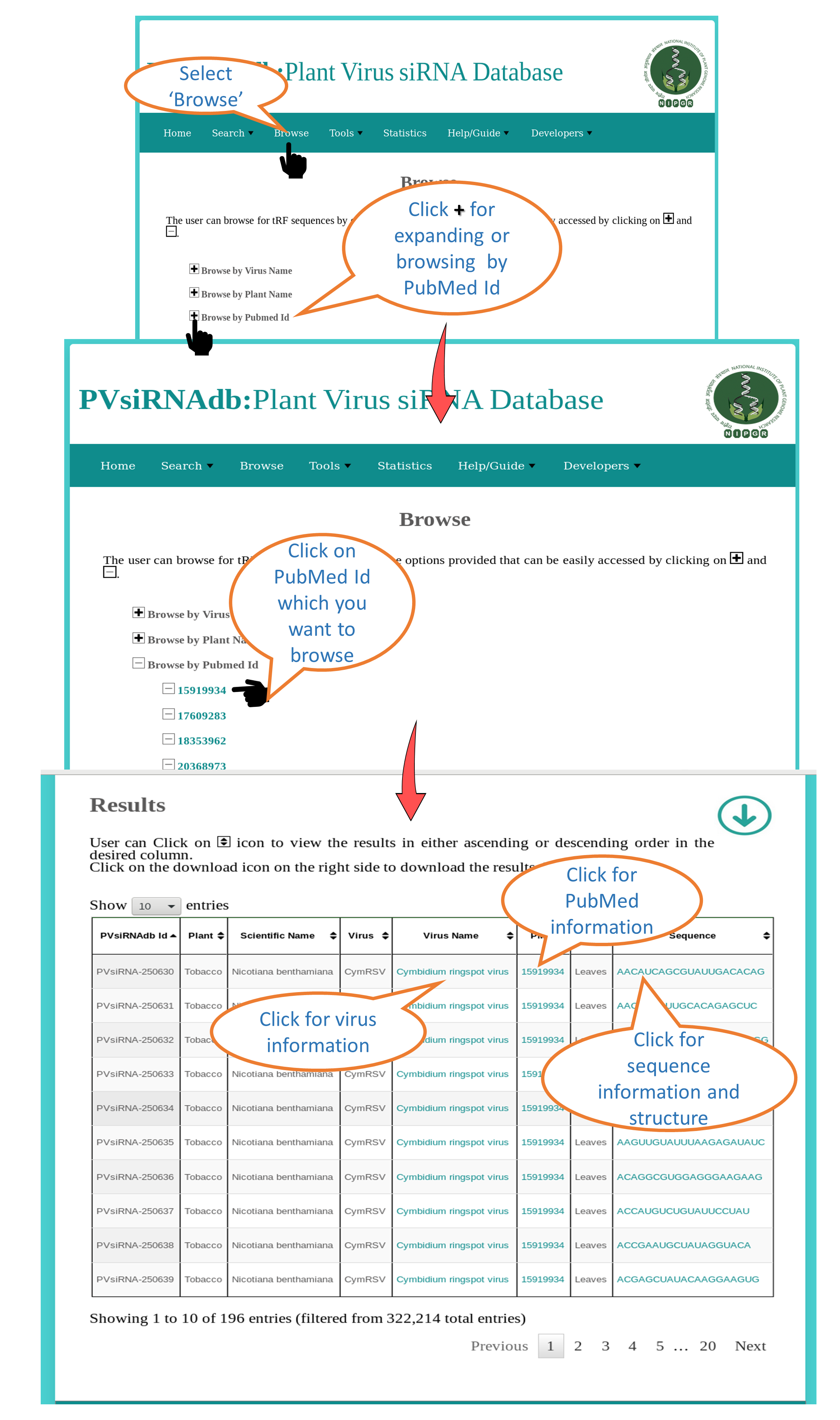
Browse by PubMed ID |
| This interface allows the user to browse all siRNAs available in PVsiRNAdb fetched from specific PubMed ID. All the available information will be dispalyed e.g. Virus, Sequence, PMID et |
Tools
BLAST |
| User can run a BLAST search for sequences. The server provides options to BLAST against a specific type of virus's siRNA or against whole PVsiRNA database. Different E-values are provided , from which user can choose one. Default E-value is set at 10 |

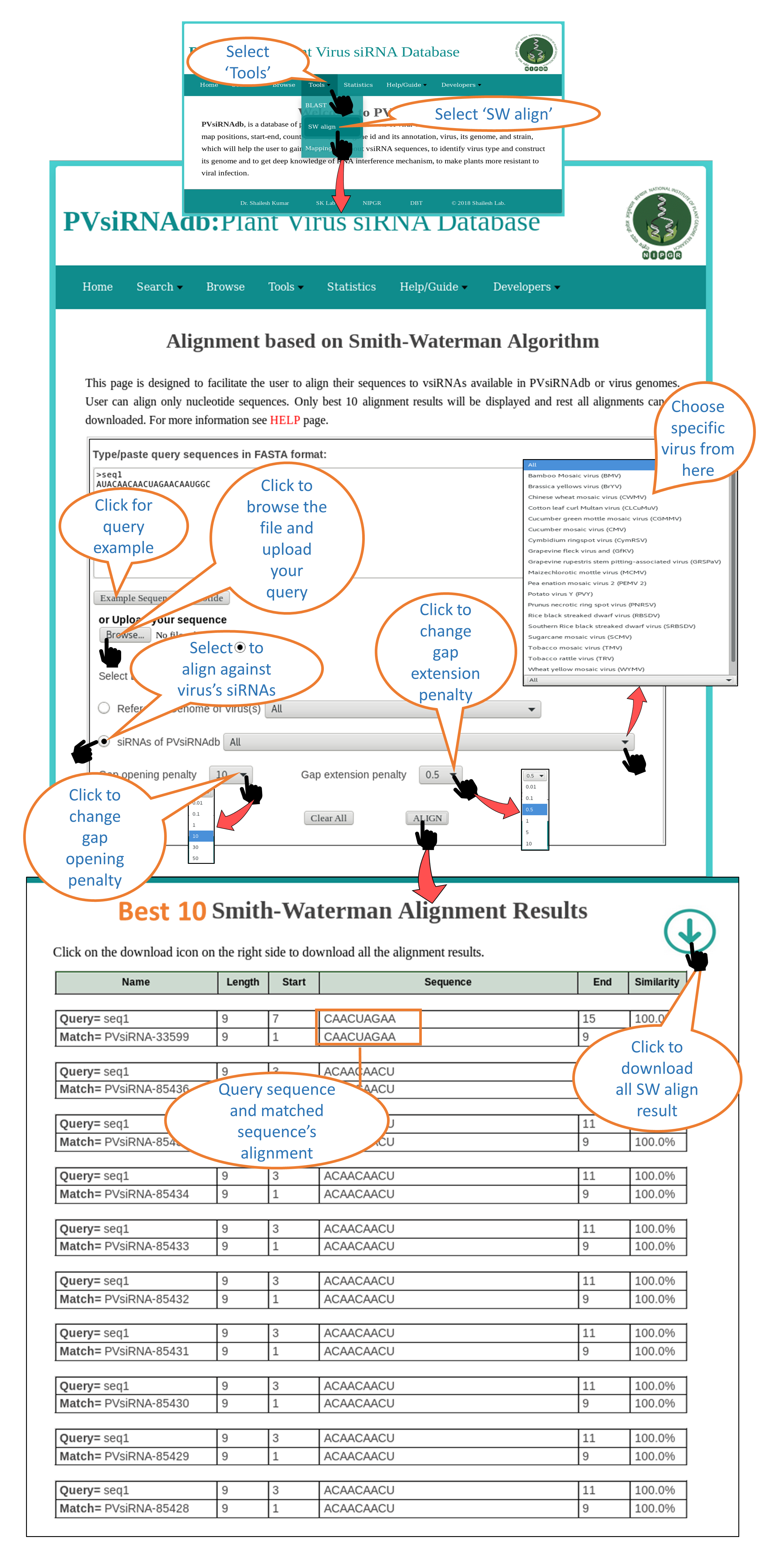
SW align |
| SW align i.e. Smith-Waterman Alignment page assists the user to run a Local alignment of a sequence using Smith-Waterman algorithm. User can align a sequence either against the reference genome of specific virus or against the reference genome of all the virus available in PvsiRNAdb OR user can align a sequence against the siRNAs of specific virus or all virus data present in PVsiRNA database. |
1. Alignment against reference genome of virus
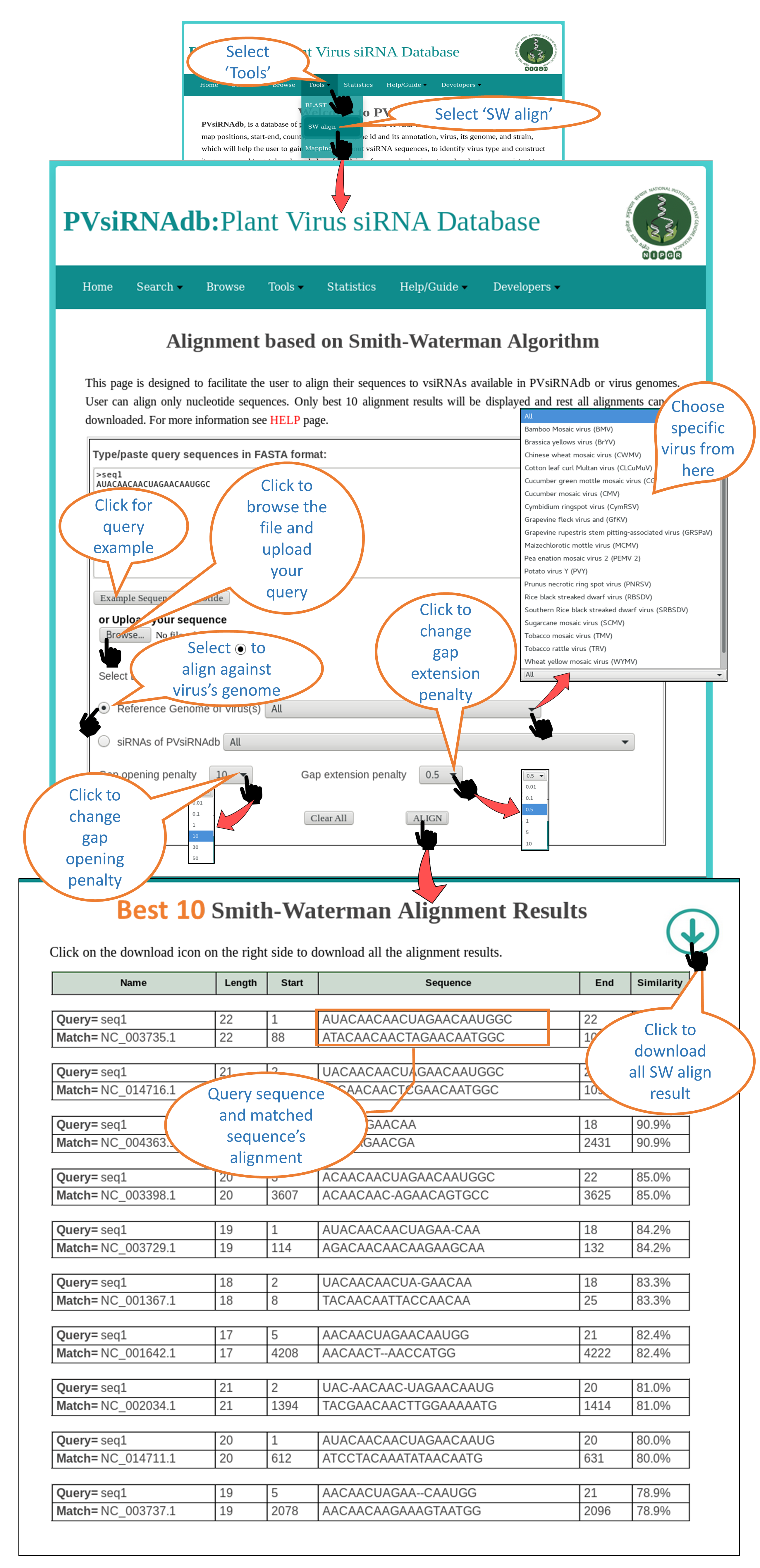
2. Alignment against siRNAs of virus
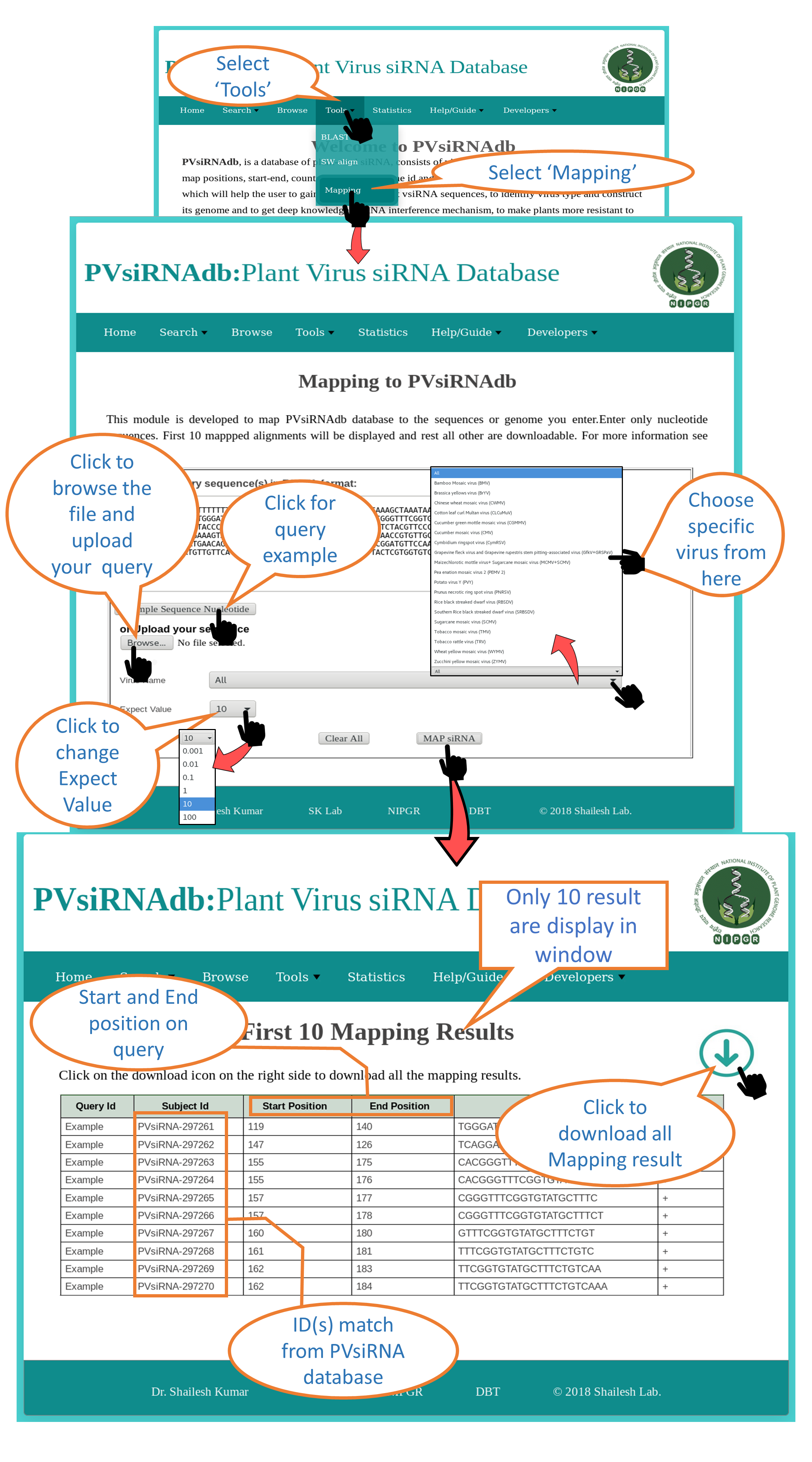
Mapping |
| This module maps the PVsiRNAdb siRNA's on the user provided nucleotide sequence. Mapping can be done against different specific virus's siRNAs or all the siRNAs available in PVsiRNAdb. Different E-value can be provided for mapping. |