Help
This page is designed to help the user to understand MedProDB and use it effectively. Here user can get the details of all the procedures present in different modules in a step-wise manner. User can directly go to the particular section by clicking the respective menu present in the main table.
Search
Global Search |
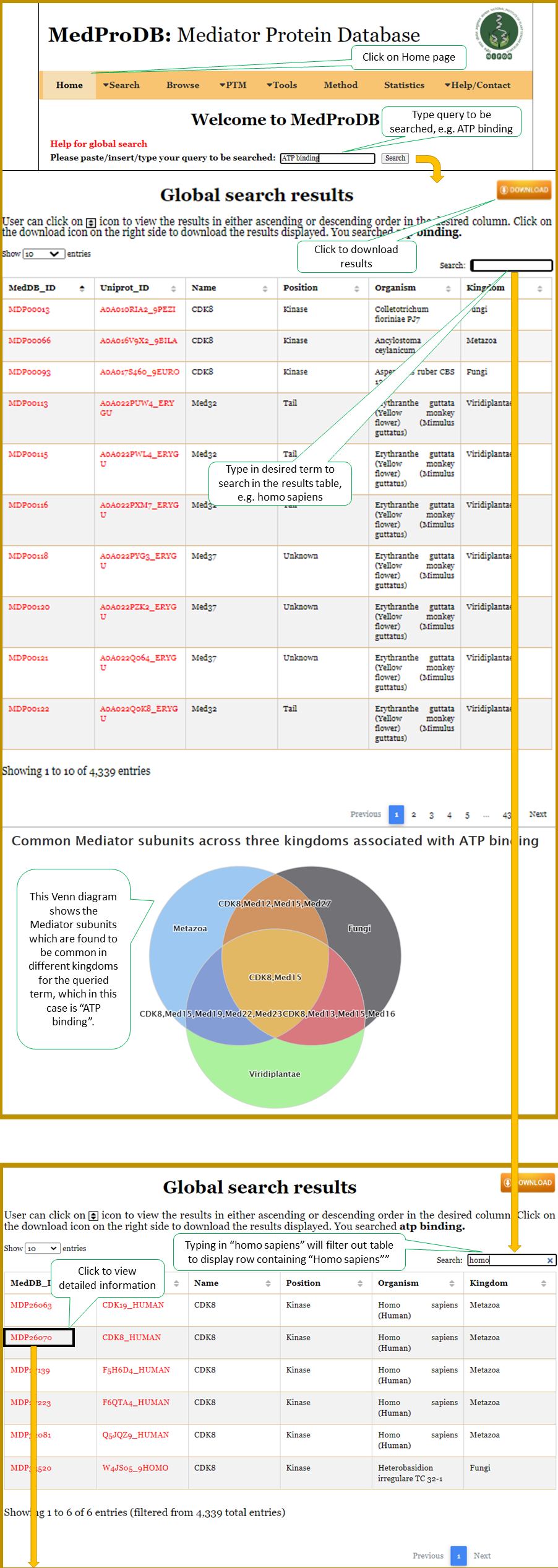
| This is a very basic and user friendly global search option. User can search any query in the text box provided (e.g. 'ATP binding' or 'transcription'). List of Mediator protein sequences will be displayed wherever the query is matched in the whole database (will be searched in function, interaction, description, disease section of the database). |
1. Global Search
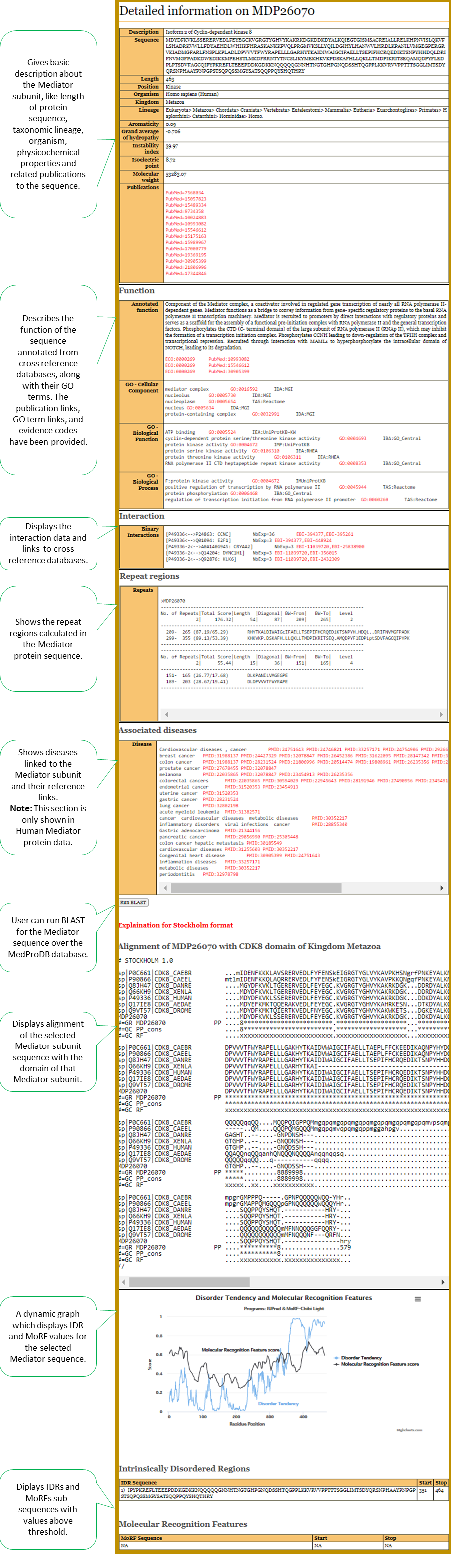
Details section of the selected Mediator sequence
Simple Search |
| This is a very basic and user friendly search option. User can search any query in any of the fields (e.g. Name Head) of the database. Results will be displayed according to the selected fields to be displayed. |
1. Default Condition
2. When clicked on "Containing"
3. When clicked on "Exact"
Advanced Search |
| This is a level-up from the usual simple search option. User can search MedProDB by using multiple queries combined with logical operators like AND / OR / != / >= / <=. All the data matching with the query, will be displayed with all other related information available in the database. |
Browse
Browse by Name |
| This module allows the user to browse all the Mediator sequences according to the mediator subunit type in MedProDB. There are 39 types of Mediator sequence data present in this database. |
Browse by Kingdom |
| This module allows the user to browse all the Mediator sequences according to the Kingdom in MedProDB. There are 3 Kingdom's data present in this database. |
Browse by Position |
| This module allows the user to browse all the Mediator sequences according to the mediator subunit position in MedProDB. There are 4 positions of Mediator subunit sequence data present in this database. |
Tools
SW Align |
| SW align i.e. Smith-Waterman Alignment page assists the user to run a Local alignment of a sequence using Smith-Waterman algorithm. User can align a protein sequence to all the Mediator protein sequences available in MedProDB. |
BLAST |
| User can run a BLAST search for protein sequences. The server provides hits against whole MedProDB. Different E-values are provided, from which user can select the suitable one. Default E-value is set at 10. |
IUPred |
| User can provide mediator protein sequence and server will predict Intrinsically Disordered Regions in the input sequence. User can select disorder types to be predicted and server generates a dynamic graphical representation of IDRs. |
MoRF |
| User can provide mediator protein sequence and server will predict Molecular Recognition Features in the input sequence. Server generates a dynamic graphical representation of MoRFs. |
Post Translational Modification sites |
| User can click on four types of PTM sites, i.e. Acetylation, Glycosylation, Methylation and Phosphorylation. Server provides user with downloadable raw data of 8 modal organisms. |