Welcome to PTPAMP
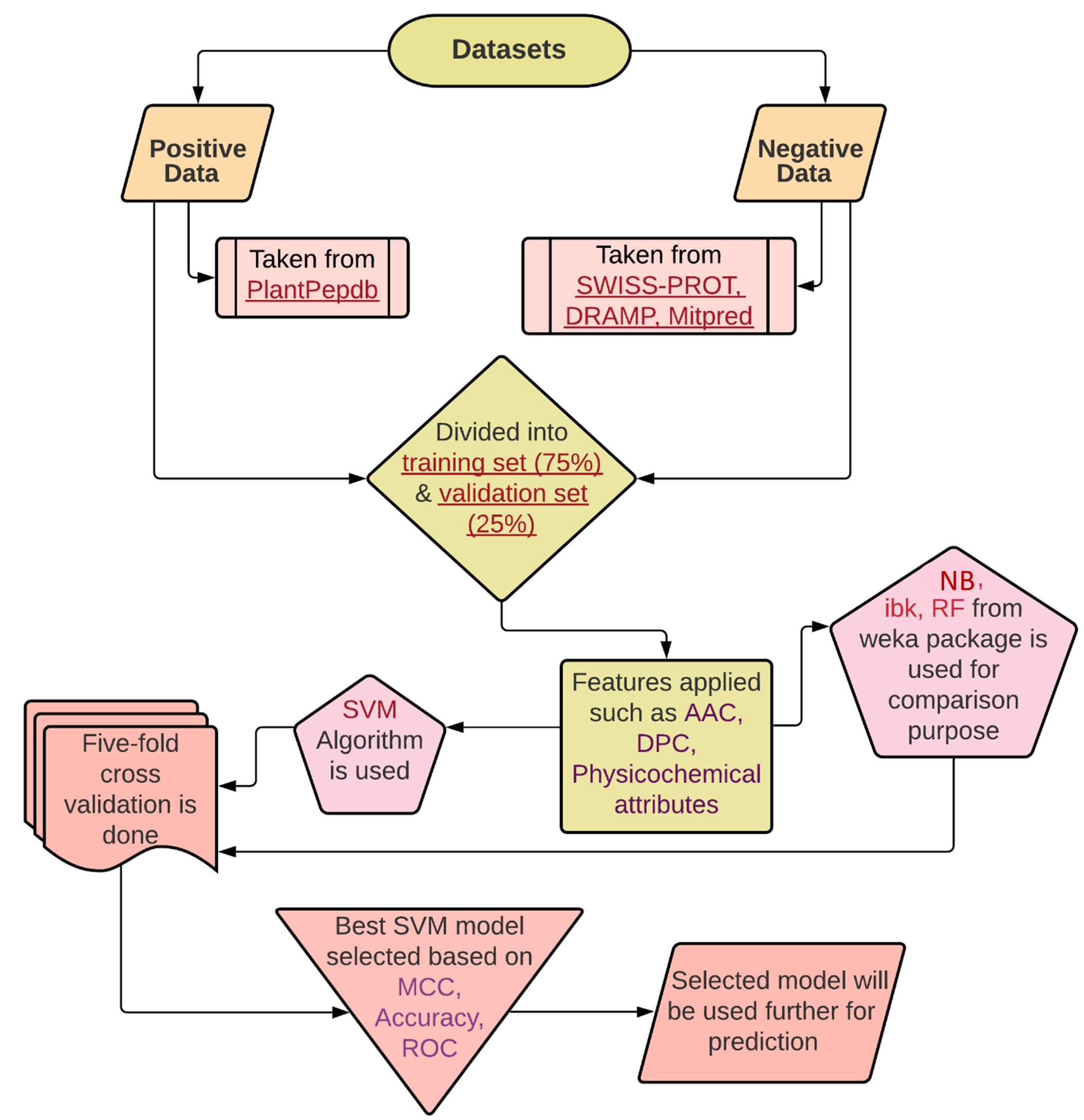
PTPAMP is a prediction tool 🔧 to predict the activity of plant-derived peptides in terms of being antimicrobial or not. It categorizes the result into four different
activities named antimicrobial, antibacterial, antifungal, and antiviral. This tool has a module for screening query sequences to visualize their probability of
being classified as each of the activities mentioned above. The classification scores of each activity is calculated by the implemented SVM models.
Features on which the PTPAMP models are based upon:
• AAC (Amino Acid Composition)
• DPC (Dipeptide Composition)
In PTPAMP, there is a module of peptide designing to mutate the query peptide in order to design certain mutants with modified properties and antimicrobial potential.
Apart from screening and designing, the protein scan module is also there to scan throughout the query protein sequences as per the sequence and overlapping
length provided by the user to find out any novel peptide with antimicrobial activity.
For more information go through the Help page.