Welcome to AlnC
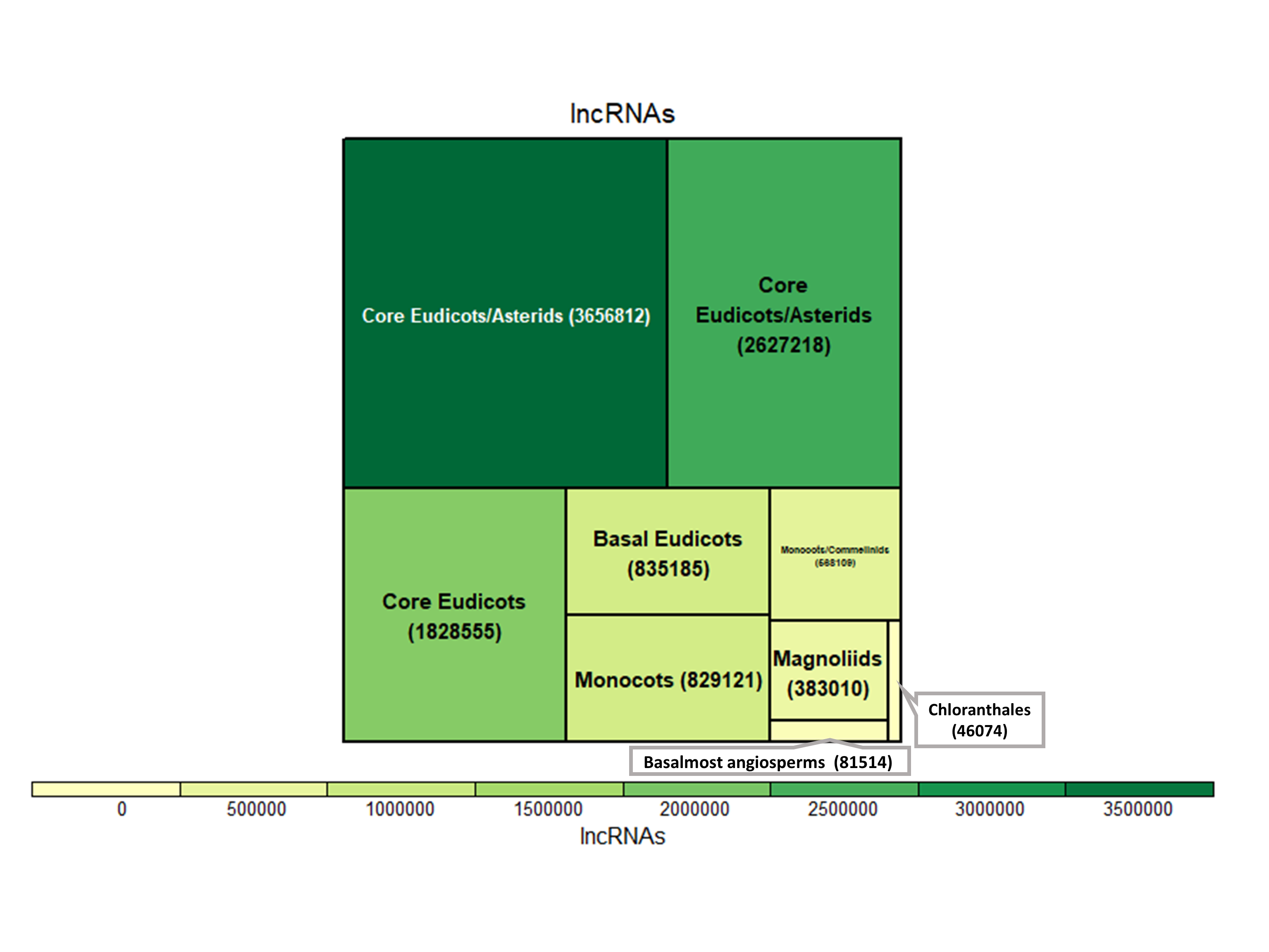
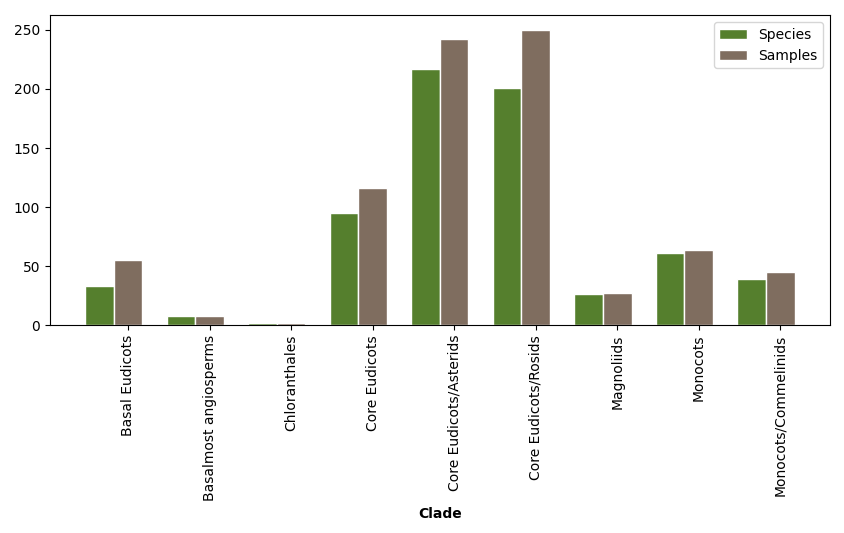
AlnC is an extensive database of long non-coding RNAs (lncRNAs) in Angiosperms. Here, we have incorporated a total of 10,855,598 lncRNAs, annotated in 809 RNA-seq samples of 682 flowering plants by using machine learning approach previously described by Singh et al., 2017. All transcriptome data available at 1KP project initiative was processed and used to annotate the lncRNAs. At AlnC, different kind of modules are available to search and download various kind (e.g. sample, tissue, sequence, structure, length etc.) of information related to lncRNAs in each plant species.
Features to AlnC include:
1. Basic lncRNA annotation from whole plant-samples to organs/tissues that can be explored on the basis of taxonomic ranks.
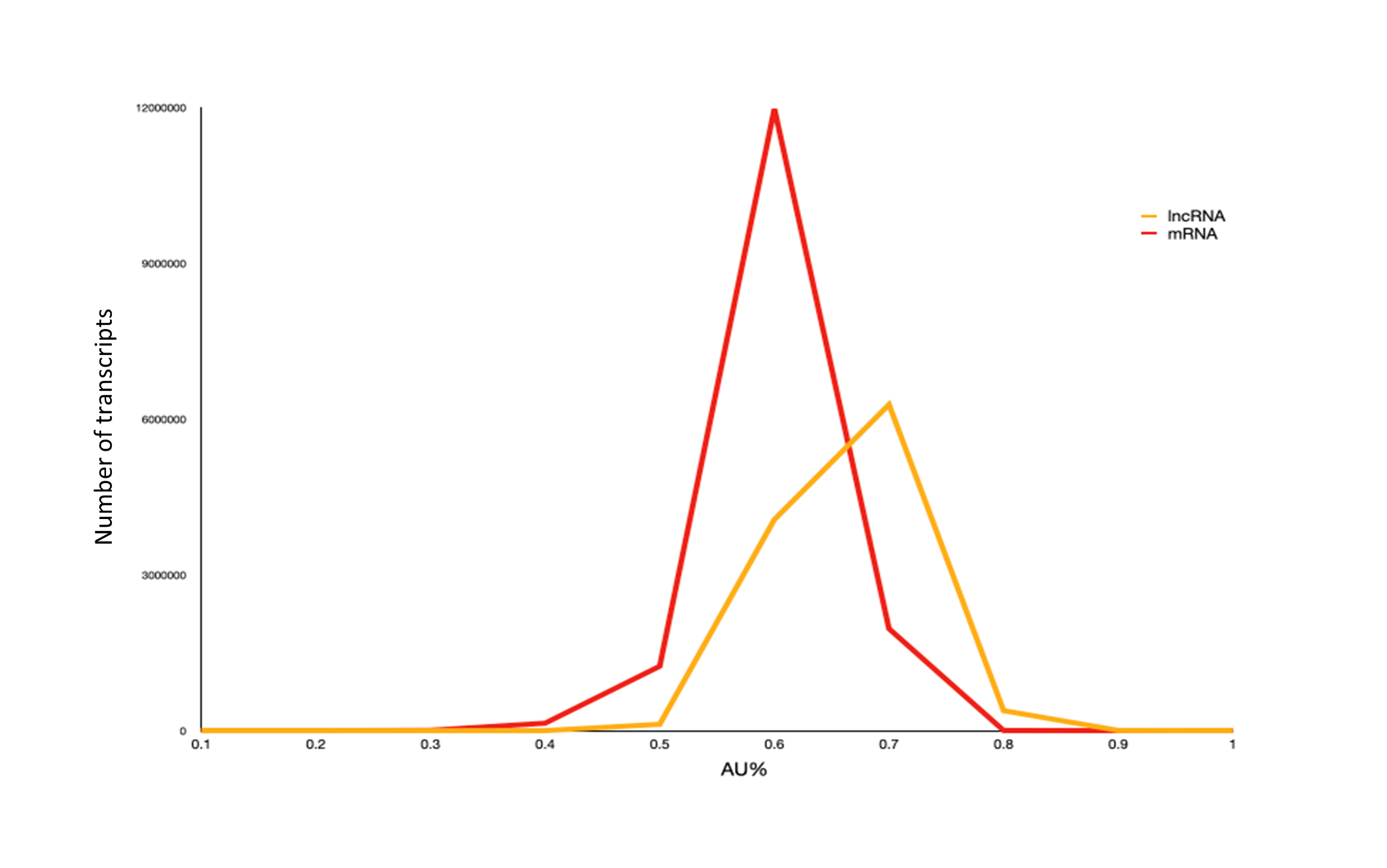
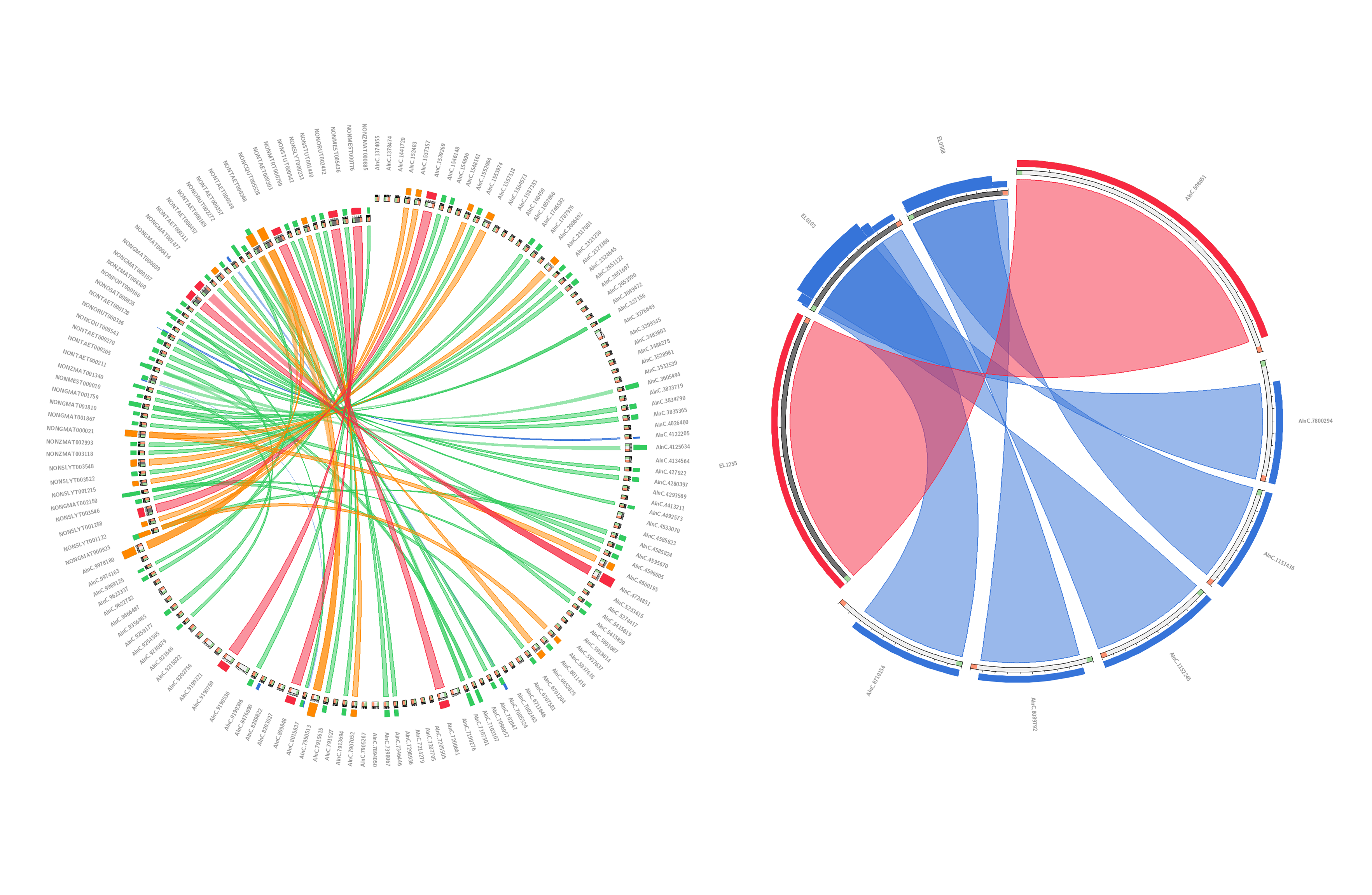
2. Each lncRNA entry is tagged with sample information, NCBI metadata, sequence and structure characteristics, and Non-coding probability score.
3. All tabular information and FASTA files of lncRNA entries as well species wise lncRNA transcript models is available for immediate download.
4. All lncRNAs are recorded with possible sORFs within its sequence.
© Developed by Dr. Shailesh Laboratory | National Institute of Plant Genome Research (NIPGR), New Delhi, India | DBT