tncRNA Toolkit
| 1. Brief Introduction | 2. tncRNA Toolkit workflow |
| 3. Download | 4. Prerequisites |
| 5. Installation | 6. Inside the Package |
| 7. Usage | 8. Output |
1. Brief Introduction
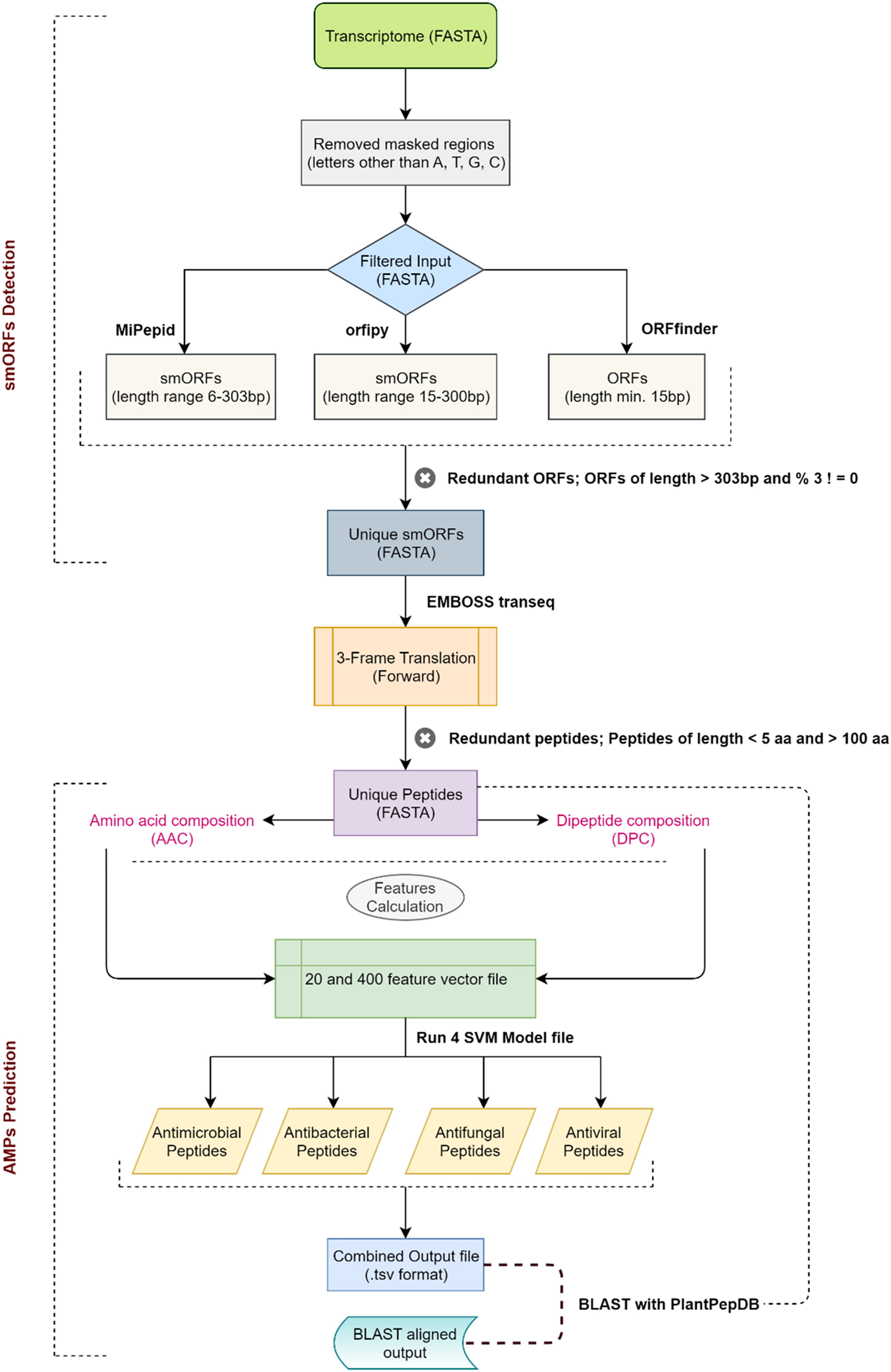
smAMPsTK Toolkit is designed for the identification of antimicrobial peptides from plant's transcriptome data. smAMPsTK detects peptides of four different activities i.e. antimicrobial, antibacterial, antifungal, and antiviral.
2. smAMPsTK Toolkit workflow
3. Download
Download the smAMPsTK Toolkit package from below link:
smAMPsTK-Toolkit.tar.gz
Note: This pipeline is tested on CentOS 7/8.
4. Prerequisites
1. python3
Python modules: orfipy (v0.0.4), orffinder (v1.8), pandas (v1.3.5), configparser (v5.2.0), biopython (v1.81), numpy (v1.21.6), scikit-learn (v 0.21.3) and scipy (v1.7.3)
Steps to make python enviroment:
conda create -n py3.7
conda activate py3.7
conda install python==3.7
Steps to create python environment for packages:
python3.7 -m venv sklearn-env (Created sklearn_env)
source sklearn_env/bin/activate (Activate sklearn_env)
pip install -U scikit-learn==0.21.3 (Installtion of sklearn version)
pip install package's name==version (Install the prerequisites)
OR
pip3 install <module name> --user
2. EMBOSS Transeq (v6.6.0)
3. ncbi-blast (v2.9.0+)
[Note: python3, EMBOSS, and ncbi-blast are needed to be globaly installed or included it in the path.]
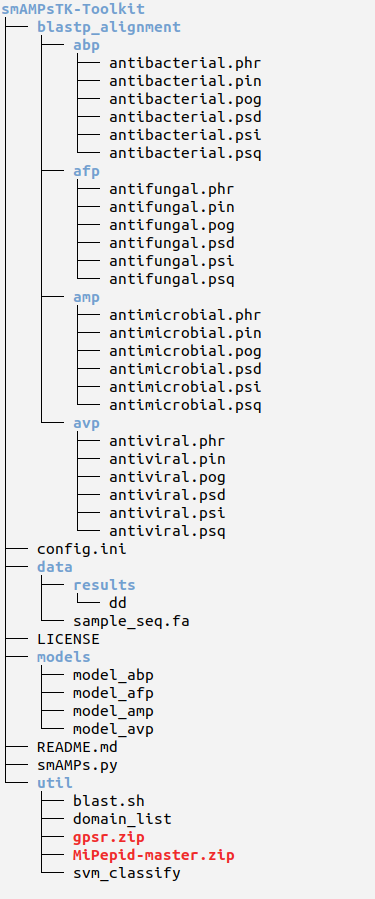
4. MiPepid tool
5. GPSR package
5. SVM classify
[Note: MiPepid tool, and GPSR package provided as zip file. SVM classify is provided as a script.]
7. InterProScan (v5.56-89.0)
5. Installation
Extract the tarball using the command:
tar -zxvf smAMPsTK-Toolkit.tar.gzcd smAMPsTK-Toolkit/Installing MiPepid and GPSR:
unzip MiPepid-master.zipunzip gpsr.zip[Note: GPSR will be used from util directory]
6. Inside the Package
This distribution includes the python3 script smAMPs.py and other scripts in the ‘util’ folder which are needed to run the main script.
7. Usage
First change the path of input file, MiPepid, InterProScan, and output folder in the provided config.ini file.
Once config file is set according to the user's path;
For AMPs prediction from smORFs:
python3.7 smAMPs.py config.ini -th 0 It will automatically create the bowtie index alongwith needed files in "lib/indexes/<provided species name>"
For domain search -domain argument is needed to be provided, like this:
python3.7 smAMPs.py config.ini -th 0 -domain
8. Output
In the provided directory with ‘-o’ option, smAMPsTK Toolkit provides 22 result files:
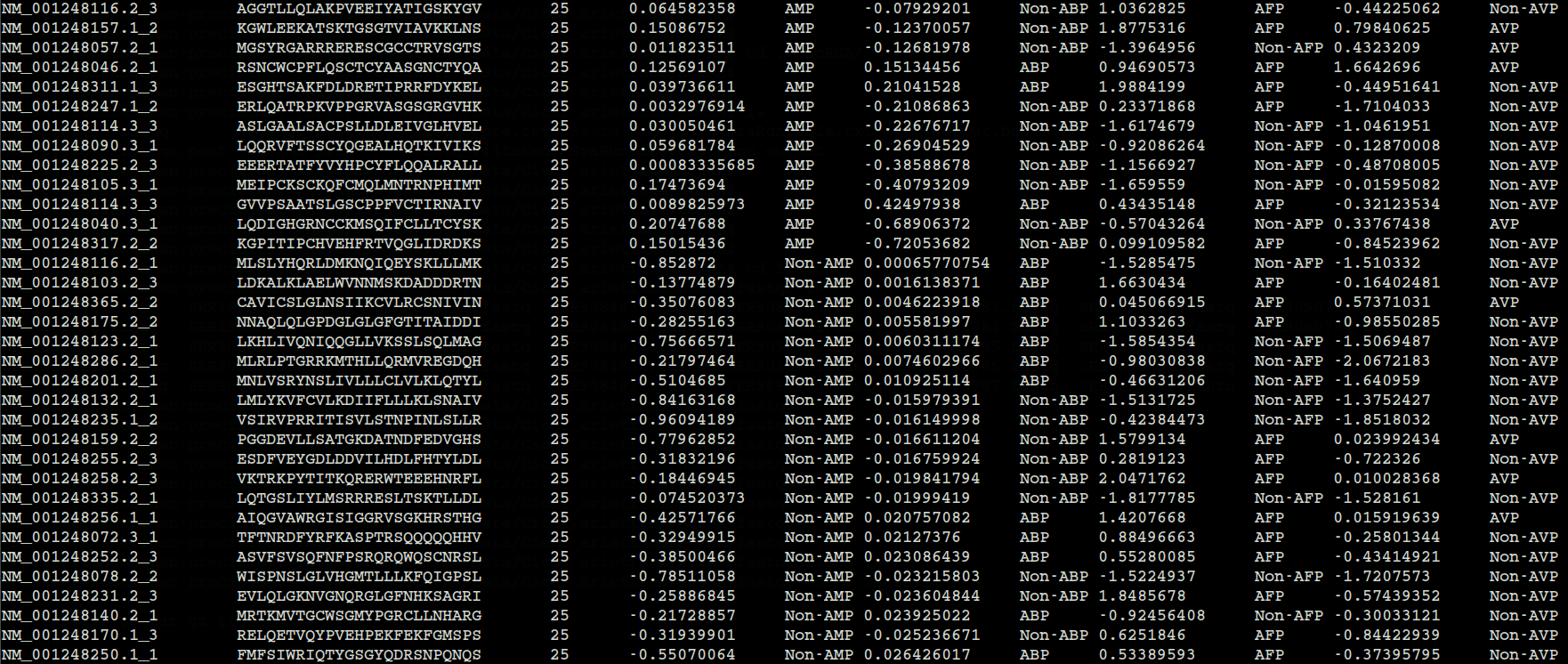
1. output.tsv
smAMPsTK detects peptides of four different activities i.e. antimicrobial, antibacterial, antifungal, and antiviral are stored in output.tsv.
Total of eleven columns in output.tsv (shown in Figure 3) are as follows:
1. Transcript ID/Identifier:
Transcript IDs/Identifier corresrponding to the mRNA sequences present in the sequence identifier.
2. Peptide Sequence:
This column contains peptides which are predicted by the MiPepid tool for the given mRNA sequences.
3. Peptide Length: Length of Peptide
4. Prediction Score: AMP (antimicrobial peptide) prediction score.
5. Predicted Label: AMP (antimicrobial peptide) predicted label.
6. Prediction Score: ABP (antibacterial peptide) prediction score.
7. Predicted Label: ABP (antibacterial peptide) predicted label.
8. Prediction Score: AFP (antifungal peptide) prediction score.
9. Predicted Label: AFP (antifungal peptide) predicted label.
10. Prediction Score: AVP (antiviral peptide) prediction score.
11. Predicted Label: AVP (antiviral peptide) predicted label.
2. Fasta Files
The smAMPsTK Toolkit categorizes peptide sequences into their predicted classes, such as antimicrobial, antibacterial, antifungal, and antiviral, and then records them in four distinct files: amp_align.fa, abp_align.fa, afp_align.fa, and avp_align.fa respectively.
In the specified directory, when using the '-domain' option and '-o' option, smAMPsTK Toolkit provides 8 additional files for domain search.
3. Domain Files
The smAMPsTK Toolkit utilizes InterProScan to analyze peptide fasta files obtained from the preceding step. For each fasta file, including amp_align.fa, abp_align.fa, afp_align.fa, and avp_align.fa, InterProScan identifies protein domains and generates files containing information about Interpro IDs and these protein domains associated with the peptides i.e. domain_result_amp, domain_result_abp, domain_result_afp, and domain_result_avp. respectively.
[Note: Above three listed files types are main output files from smAMPsTK Toolkit rest are the intermediate files i.e. all_forf.txt, out.txt, pep_f.dpc, pep_seq_f.txt, all_orf.tab, input.fa, pep_f_col.aac, pep_f.mono, pep_seq.tab, abp_res, afp_res, all_orf.txt, amp_res, avp_res, pep_f_col.dpc, pep_f.sfa, and pep_seq.txt]
