Search
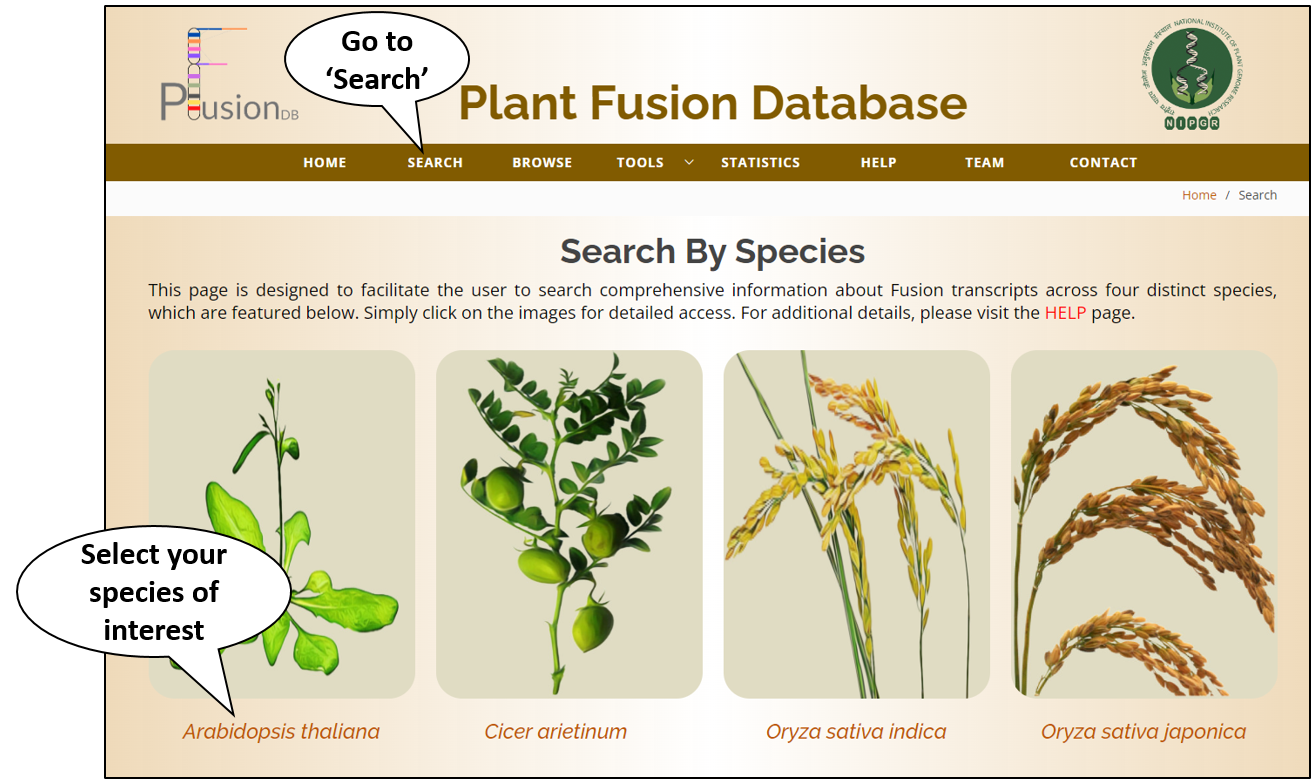
Simple Search |
| This is a very basic and user friendly search option. User can search any query in any of the fields (e.g. Ensembl ID AT2G34480) of the database. Results will be displayed according to the selected fields to be dispalyed. e.g. PFusion ID, Fusion Name, Gene Name, Breakpoint position, Pattern, Fusion Type, Tissue, Treatment, Tool, and SRR ID etc. |

Advanced Search |
| This is a level-up from the usual simple search option. User can search PFusionDB by using multiple queries combined with logical operators like AND / OR / NOT. All the data matching with the query, will be displayed with all other related information available in the database. |

Browse
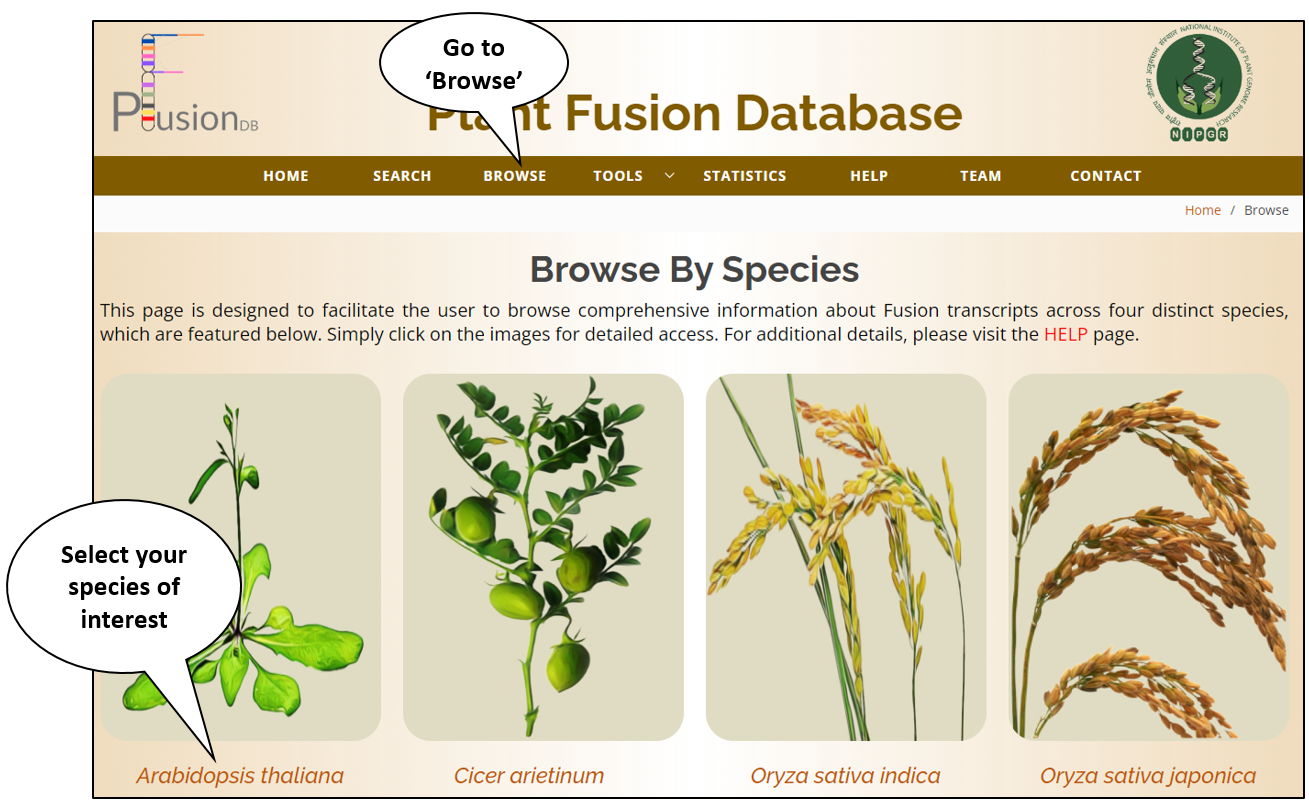
Browse by Fusion Type |
| This module allows the user to browse all the fusion transcripts according to the fusion type in PFusionDB. Fusion transcripts are classified into two types based on their chromosomal origin: InterType and IntraType. Further IntraType is categorized into two classes based on the distance between the two genes involved in fusion formation: Proximal type and Distal type. All available information related to the selected fusion type will be dispalyed e.g. PFusion ID, Fusion Name, Gene Name, Breakpoint position, Pattern, Fusion Type, Tissue, Treatment, Tool, and SRR ID etc. |

Browse by Tissue Type |
| This module allows the user to browse all the fusion transcripts according to tissue type. All available information related to the selected tissue will be dispalyed e.g. PFusion ID, Fusion Name, Gene Name, Breakpoint position, Pattern, Fusion Type, Tissue, Treatment, Tool, and SRR ID etc. |

Browse by Tools |
| This module allows the user to browse all fusion transcripts analyzed by the five different tools: EricScript, MapSplice, Squid, STAR-Fusion and TrinityFusion. All available information related to the selected tool will be dispalyed e.g. PFusion ID, Fusion Name, Gene Name, Breakpoint position, Pattern, Fusion Type, Tissue, Treatment, Tool, and SRR ID etc. |

Browse by Chromosome |
| This module allows the user to browse all fusion transcripts by their Chromosome number. All available information related to the selected chromosome will be dispalyed e.g. PFusion ID, Fusion Name, Gene Name, Breakpoint position, Pattern, Fusion Type, Tissue, Treatment, Tool, and SRR ID etc. |

Browse by Pattern |
| This module allows the user to browse all fusion transcripts by their breakpoint pattern. All available information related to the selected breakpoint pattern will be dispalyed e.g. PFusion ID, Fusion Name, Gene Name, Breakpoint position, Pattern, Fusion Type, Tissue, Treatment, Tool, and SRR ID etc. |

Browse by Treatment Type |
| This module allows the user to browse all fusion transcripts by their treatment type. All available information related to the selected treatment will be dispalyed e.g. PFusion ID, Fusion Name, Gene Name, Breakpoint position, Pattern, Fusion Type, Tissue, Treatment, Tool, and SRR ID etc. |

Browse by Frequency |
| This module shows the PFusion ID of the highest occuring fusions under three categories namely tissue-wise, fusion-type wise and condition wise. The user can select any of the three categories to view the top occuring PFusion IDs. Further, the user can click on any of the PFusion ID to view detailed information of all the occurences of that fusion transcript. All available information related to the selected fusion transcripts will be dispalyed e.g. PFusion ID, Fusion Name, Gene Name, Breakpoint position, Pattern, Fusion Type, Tissue, Treatment, Tool, and SRR ID etc. |

Tools
BLAST |
| User can run a BLAST search for nucleotide sequences. The server provides hits against whole PFusionDB database. Different E-values are provided, from which user can select the suitable one. Default E-value is set at 10. |

SW Align |
| SW align i.e. Smith-Waterman Alignment page assists the user to run a local alignment of a sequence using Smith-Waterman algorithm. User can align a nucleotide sequence to all the fusion transcript junction sequences available in PFusionDB. |

Mapping |
| This module maps the fusion transcript junction-sequences that are available in PFusionDB on the user provided nucleotide sequence. Mapping can be done against all the junction sequences available in PFusionDB. User can select different E-values before mapping. Default E-value is set at 10. |